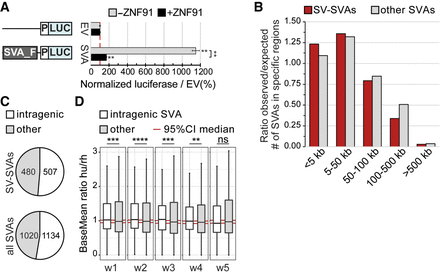
SV-SVAs reside in gene-regulatory regions. (A) Luciferase activity of construct without (EV) and with an SVA element (SVA_F) upstream of a minimal promoter (P) in mESCs. N3n9, two-sided t-test with Bonferroni correction, (**) P < 0.01. Error bars: SEM. (B) Distribution of SV-SVAs (red) and non-SV-SVAs (gray) per distance to TSS. Only SVAs > 1 kb are shown. (C) Number of SV-SVAs and non-SV-SVAs that are intragenic is comparable (χ2(1, N = 2154) = 1.10, P = 0.29). Only SVAs > 1000 bp are shown. (D) Box plots showing base mean expression ratio (human/rhesus) for transcripts with an intragenic SVA in humans (white; 1151) and without (gray; 23,296) in ESC-derived cortical organoids of 1- to 5-wk old. Red line shows 95% CI of 10,000× bootstrapped median of transcripts without an SVA with sample size of 1151. Wilcoxon rank-sum test: (****) P < 0.0001, (***) P < 0.001, (**) P < 0.01, ns = not significant.











