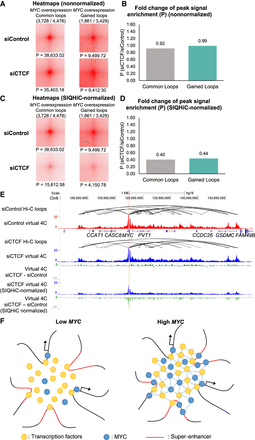
Chromatin contacts can be disrupted through CTCF siRNA knockdown. (A) siControl and siCTCF Aggregate Peak Analysis plots at 5-kb resolution showing the nonnormalized aggregate signal at “Common” and “Gained” chromatin loops previously identified in High MYC cells. (P) Peak signal at the center pixel. (B) Fold change of nonnormalized peak signal enrichment (P) at common and gained loops between siControl and siCTCF cells. (C) Same APA analysis as in A but with SIQHiC normalization applied. (D) Fold change of SIQHiC-normalized peak signal enrichment (P) at common and gained loops between siControl and siCTCF cells. (E) Genome browser view showing siControl and siCTCF virtual 4C signal of interactions at the MYC promoter (red and blue tracks, respectively). The difference between the siControl and siCTCF virtual 4C signals are shown with and without SIQHiC normalization (green tracks). (F) Proposed model of overexpressed MYC accumulating at canonical promoter binding sites, binding to spatially adjacent super-enhancers, and forming protein–protein interactions with other transcription factors to stabilize chromatin interactions within the domain.











