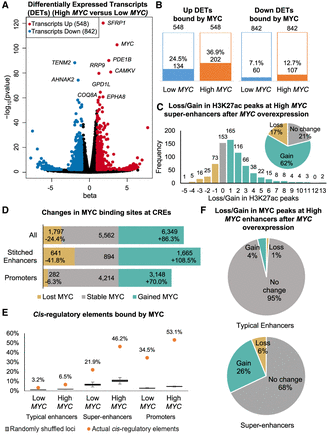
MYC overexpression leads to differential transcript expression, differential H3K27ac signal, and increased MYC binding at super-enhancers. (A) Significantly up-regulated (red) and down-regulated (blue) transcripts after MYC overexpression (|beta| > 1, FDR < 0.05, Wald test). Top 10 significantly regulated transcripts are labeled. (B) Number of differentially expressed transcripts (DETs) bound by MYC in Low MYC and High MYC cells. (C) Bar graph and pie chart showing gain and loss of H3K27ac peaks at super-enhancers after MYC overexpression. (D) Bar graph showing gain and loss of MYC binding sites at stitched enhancers and promoters after MYC overexpression. (E) Proportion of typical enhancers, super-enhancers, and promoters being bound by MYC compared to randomly shuffled coordinates. The proportion of MYC-bound CREs are shown as orange dots. Box plots show 1000 iterations of MYC occupancy at random genomic loci of the same size and on the same chromosome as the actual CREs. (F) Pie charts showing gain and loss of MYC ChIP-seq peaks at typical enhancers (top) and super-enhancers (bottom) after MYC overexpression.











