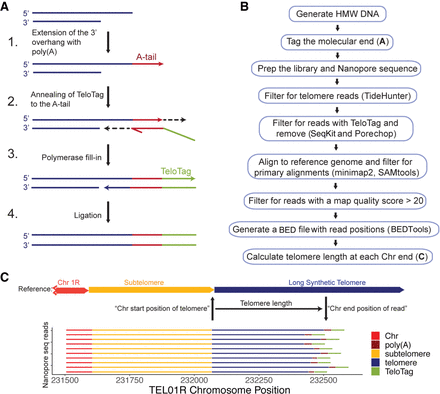
Telomere length measurement by nanopore sequencing. (A) Schematic of method to tag the molecular end of the chromosome. (1) Terminal transferase adds poly(A) sequence to the 3′ overhang of the telomere. (2) A complementary oligo(dT) sequence on the TeloTag oligonucleotide end containing a unique TeloTag sequence is annealed to the poly(A) sequence. (3) Polymerase fill-in generates double-stranded blunt ends. (4) Ligase is used to seal the nick. Nanopore adaptors are then added using the manufacturer's protocol. (B) Bioinformatic pipeline to generate telomere length information (see Methods). (C) Telomere length is calculated using the chromosomal position of each telomere sequence start and calculating the distance to the TeloTag sequence at the chromosome end. A random sample of 10 sequence reads aligned at TEL01R is presented as an example.











