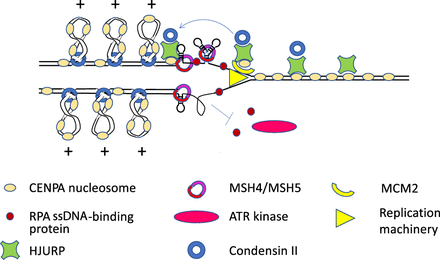
Speculative model of replication through α-satellite. Holliday junction recognition protein (HJURP) associates with CENPA nucleosomes before S-phase and recruits the condensin II complex. At the replication fork, HJURP and the MCM2 subunit of the replication machinery work together to assure that CENPA nucleosomes reassemble behind the fork. DNA secondary structures form on single-stranded repetitive DNA behind the fork, and HJURP and mismatch repair proteins (MSH4 and MSH5 are shown) bind to them and resolve them. Condensin II complexes extrude positively supercoiled DNA loops, and the positive torsion inhibits the binding of replication protein A (RPA), which binds single-stranded DNA and must accumulate in order for the ATR serine/threonine kinase (ATR) to signal that DNA damage has occurred and to arrest replication. This inhibition by condensin II allows time for secondary structures to be resolved. Condensin II is also needed with HJURP to assemble new CENPA nucleosomes in G1, and condensin-mediated loops may play a role in the organization of the kinetochore.











