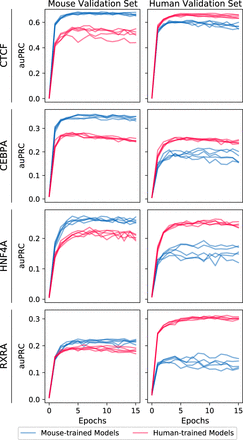
Model performance over the course of training, evaluated on held-out validation data from mouse (left) and human (right) Chromosome 1. Five models were independently trained for each TF and source species (mouse-trained models in blue, human-trained models in red). Values at epoch 0 are evaluations of models after weight initialization but before training (akin to a random baseline). Note that auPRCs are not directly comparable between different validation sets because ground truth labels are derived from a different experiment for each data set; the area under the precision-recall curve (auPRC) will depend on the fraction of sites labeled as bound as well as model prediction correctness.











