Whole-genome resequencing of temporally stratified samples reveals substantial loss of haplotype diversity in the highly inbred Scandinavian wolf population
- Agnese Viluma1,
- Øystein Flagstad2,
- Mikael Åkesson3,
- Camilla Wikenros3,
- Håkan Sand3,
- Petter Wabakken4 and
- Hans Ellegren1
- 1Department of Evolutionary Biology, Uppsala University, SE-752 36 Uppsala, Sweden;
- 2Norwegian Institute for Nature Research (NINA), Torgard, NO-7485, Trondheim, Norway;
- 3Department of Ecology, Swedish University of Agricultural Sciences, SE-739 93 Riddarhyttan, Sweden;
- 4Faculty of Applied Ecology, Agricultural Sciences and Biotechnology, Inland Norway, University of Applied Sciences, Campus Evenstad, NO-2480 Koppang, Norway
Abstract
Genetic drift can dramatically change allele frequencies in small populations and lead to reduced levels of genetic diversity, including loss of segregating variants. However, there is a shortage of quantitative studies of how genetic diversity changes over time in natural populations, especially on genome-wide scales. Here, we analyzed whole-genome sequences from 76 wolves of a highly inbred Scandinavian population, founded by only one female and two males, sampled over a period of 30 yr. We obtained chromosome-level haplotypes of all three founders and found that 10%–24% of their diploid genomes had become lost after about 20 yr of inbreeding (which approximately corresponds to five generations). Lost haplotypes spanned large genomic regions, as expected from the amount of recombination during this limited time period. Altogether, 160,000 SNP alleles became lost from the population, which may include adaptive variants as well as wild-type alleles masking recessively deleterious alleles. Although not sampled, we could indirectly infer that the two male founders had megabase-sized runs of homozygosity and that all three founders showed significant haplotype sharing, meaning that there were on average only 4.2 unique haplotypes in the six copies of each autosome that the founders brought into the population. This violates the assumption of unrelated founder haplotypes often made in conservation and management of endangered species. Our study provides a novel view of how whole-genome resequencing of temporally stratified samples can be used to visualize and directly quantify the consequences of genetic drift in a small inbred population.
Genetic diversity is a key component for long-term viability of populations in a changing environment (Lande and Shannon 1996; Lacy 1997; Saccheri et al. 1998; Reed and Frankham 2003; Sommer 2005; Lai et al. 2019). When the size of a population decreases, the maintenance of genetic diversity becomes challenging. In small populations genetic drift (random sampling of alleles) and inbreeding (mating of closely related individuals) will tend to erode genetic diversity. Although drift has a direct effect on allele frequencies in a population, inbreeding increases the frequency of homozygotes, which in turn reduces the effective population size and effective frequency of recombination (Charlesworth 2003). This may lead to the accumulation of recessive deleterious alleles across the genome (Charlesworth and Charlesworth 1999; Rogers and Slatkin 2017) and the associated risk for inbreeding depression (Charlesworth and Willis 2009; Hedrick and Garcia-Dorado 2016).
There is a well-established theoretical framework for the study of inbreeding and genetic drift and how they contribute to the loss of genetic diversity (Wright 1931). Empirically, loss of genetic diversity may be indirectly estimated by analyzing pedigree information (Lacy 1997; Grueber and Jamieson 2008; Jansson and Laikre 2014), although this is limited to the few populations for which such information is available. Many conservation genetic studies have quantified genetic diversity in populations using molecular analyses, now feasible on a genome-wide scale (e.g., Prado-Martinez et al. 2013; Abascal et al. 2016; Kardos et al. 2018). Typically, these studies provide a snapshot on contemporary levels of diversity in a population, which in itself does not easily translate into the conservation status of populations (Ellegren et al. 1993; Dobrynin et al. 2015; Díez-del-Molino et al. 2018). Moreover, monitoring actual loss of genetic diversity requires temporal studies including analyses of change in genomic parameters such as heterozygosity and inbreeding coefficient (Díez-del-Molino et al. 2018). Temporal data may not be easy to collect from natural populations and studies on genetic drift therefore tend to be restricted to model organisms (Nené et al. 2018; Subramanian 2018; Ørsted et al. 2019) and museum collections (Díez-del-Molino et al. 2018; Ewart et al. 2019; Turvey et al. 2019).
A direct but largely untested approach to study genomic erosion in a population is to follow the survival of individual haplotypes over time. The Scandinavian gray wolf (Canis lupus) population provides an excellent opportunity for this kind of study. After being widely distributed across Europe up until modern times, wolves were eradicated by human persecution, including in Scandinavia (Haglund 1968; Wabakken et al. 2001; Hindrikson et al. 2017; Wolf and Ripple 2017). After functional extinction in the late 1960s, a wolf population was reestablished in Scandinavia by breeding three immigrant founders: a pair in 1983, and a second male in 1991 (Wabakken et al. 2001; Vilà et al. 2003). The small number of founders and absence of gene flow from neighboring populations resulted in rapid increase of inbreeding (Vilà et al. 2003; Liberg et al. 2005; Åkesson et al. 2016). However, the population size increased and is currently about 480 individuals, including additional immigrants that recently have contributed to reproduction (Åkesson et al. 2016; Svensson et al. 2021).
We have shown previously that individuals of this population have accumulated long runs of homozygosity, some being inbred to an extent that entire chromosome pairs are identical by descent (Kardos et al. 2018). Here, we use whole-genome resequencing data of 76 Scandinavian wolves sampled over a period of 30 yr after the reestablishment to directly quantify tempo of genomic erosion in terms of haplotype and allele loss. Specifically, by deriving phased chromosome-level haplotypes of the founders and following their fate over time, we provide a novel empirical insight into how founder relatedness and rapid loss of large founder haplotype segments facilitates the observed high inbreeding level of the population.
Results
Diversity of the founder haplotypes
We analyzed whole-genome sequence data from 76 Scandinavian wolves (mean coverage = 27×) including the female founder of the population. By two-step statistical phasing of 107,576 SNP markers evenly spaced across the entire genome, we obtained individual haplotypes for 2333 nonoverlapping 1-Mb windows and chromosome-level haplotype information for all 38 autosomes and the X Chromosome (Supplemental File S1). Haplotype data from individuals born in 1983–1993 were used to infer haplotypes of the two unsampled male founders (Supplemental Fig. S1).
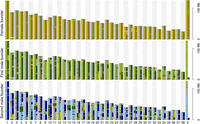
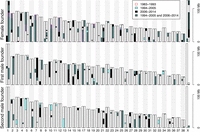
The female founder was highly heterozygous with only 84 homozygous 1-Mb windows (4% of the analyzed 2333 diploid female windows). Both male founders showed considerably higher haplotype homozygosity with 505 and 517 homozygous windows, respectively (23% of the 2217 analyzed diploid male windows in both cases) (Supplemental Table S1). Homozygous 1-Mb windows were often clustered in large blocks, in both male founders exceeding several tens of megabases, forming very long runs of homozygosity (ROH) (Supplemental Fig. S2). In addition, there was considerable haplotype sharing among the founders (Fig. 1). Specifically, considering diploid genomes, 515 1-Mb haplotypes (12% of the total number of analyzed haplotypes in the diploid genome) of the first male founder and 522 haplotypes (12%) of the second male founder were also present in the female founder. Furthermore, 839 1-Mb haplotypes (19%) of the first male founder were present in the second male founder. Pairwise comparisons of all six founder haplotypes are shown in Supplemental Table S2.
Genomic overview of the shared 1-Mb haplotypes of the founder wolves. Each haplotype contributed by a founder is assigned an individual color—two homologous haplotypes of the female founder light and dark yellow, first male founder light and dark green, and second male founder light and dark blue. To highlight identical 1-Mb windows among all six founder haplotypes, colors were assigned in hierarchical order: light yellow, dark yellow, light green, dark green, light blue, dark blue. For example, all 1-Mb windows of the dark yellow haplotype identical to those of the light yellow haplotype were colored light yellow. Similarly, all 1-Mb haplotypes of the light green haplotype of the first male founder identical to dark yellow haplotype remained dark yellow, and those identical to light yellow remained light yellow.
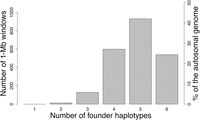
The relatively high degree of haplotype homozygosity in the male founders and significant haplotype sharing among all founders meant that only 24% of autosomal windows showed the maximally possible six different haplotypes in the three founders (Fig. 2; Supplemental Fig. S3). For 94% of windows there were four or more haplotypes, whereas <1% had only two haplotypes. On average there were 4.8 unique haplotypes per autosomal window. For the non-recombining part of the X Chromosome, 60% of windows showed the maximally possible four different haplotypes (as given by one female and two male founders), 36% showed three, and 3% only two different haplotypes. None of the 1-Mb windows was fixed for the same haplotype in all three founders.
Absence of phased 1-Mb haplotypes
We estimated the loss of genomic diversity over time by scoring the presence/absence of 1-Mb founder haplotypes in samples of the Scandinavian wolf population from three time periods. Given the genetic similarity among founders, there was some difficulty to discern whether a particular copy of a shared founder haplotype had been lost (another copy may mask its absence). To overcome this issue to some extent, we took advantage of the phased chromosome-level haplotypes and known pedigree information; linkage to the closest informative window was used to tentatively trace the origin of identical 1-Mb haplotypes.
The most detailed estimate of haplotype loss over time can be obtained for the female founder for which we had direct information on her haplotype composition across the genome. Of her 4666 1-Mb haplotypes (2 × 2333 windows), 3% were not detected in 1983–1993, 19% in 1994–2005, and 24% in 2006–2014. Haplotypes of the female founder not seen in 1983–1993 represent chromosome segments that never entered the population or that were only transmitted to unsampled offspring and then became lost (but see below). For the two founder males we could only estimate haplotype loss in the time periods 1994–2005 and 2006–2014 because data from 1983–1993 were used to indirectly infer their haplotypes. Of the initial first and second male founder 1-Mb haplotype contribution, 10% and 8%, respectively, were absent in 1994–2005, whereas 16% and 11% were absent in 2006–2014. Per-chromosome results of absent 1-Mb haplotypes are provided in Supplemental Table S3.
A particular haplotype may remain unsampled in a time period yet still be present in the population, especially if segregating at low frequency. This would lead to an overestimation of haplotype loss in that time period. For the female founder this bias appeared low because only 3% of haplotypes absent in 1983–1993 (all from a cluster from Chromosome 25) were detected in 1994–2005 and/or 2006–2014, and only 2% of haplotypes absent in 1994–2005 were observed in 2006–2014 (Supplemental Table S4). Similarly, for the first male founder 6% of haplotypes not detected in 1994–2005 were present in the sample from 2006–2014, whereas for the second male founder 32% of haplotypes not detected in the former time period were seen in the latter. However, it seems unlikely that a haplotype of the second male founder not detected in either 1994–2005 or 2006–2014 would still be present (at appreciable frequency) in the population.
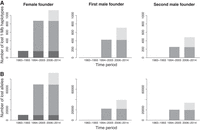
The accumulation of lost haplotypes over time is shown in Figure 3A. In 1994–2005, loss of haplotype diversity was more pronounced for the first founder couple than that of the second male founder. In 2006–2014, the number of lost 1-Mb haplotypes was similar among all three founders and comparable with the amount of loss from the second male founder in 1994–2005. Frequencies of founder haplotypes that remained present in 1994–2005 and 2006–2014 are shown in Supplemental Figure S4.
Temporal accumulation of lost founder diversity in the Scandinavian wolf population for 1-Mb haplotypes (A) and individual SNP alleles (B). Diversity lost in 1983–1993 is shown in dark gray, 1994–2005 in gray, and 2006–2014 in light gray. Only lost diversity that remained absent in later time period(s) is included.
Absence analysis of individual SNP alleles
To quantify loss of genetic diversity over time with an alternative approach not relying on statistical phasing, we followed the survival of individual alleles. Here, we used the whole set of 1,479,905 SNPs segregating in the Scandinavian population after discarding those with missing genotypes in the 1983–1993 sample. The female founder was heterozygous at 48% of these sites (Table 1). At the remaining sites (homozygous in the female founder), the other allele segregating in the population must have been contributed by one or both of the male founders. By comparing the genotypes of 1983–1993 offspring of the two male founders, we could assign the other allele for most of the female-homozygous loci. Specifically, 405,145 “female-absent” alleles were common to both male founders, whereas 104,877 alleles were unique to the first male founder and 258,191 to the second male (Table 1). The remaining 3444 alleles appeared in the samples only after 1993 and could not be assigned to a particular founder.
Initial founder contribution and gradual allele loss in the Scandinavian wolf population
Considering the genetic contribution of the female founder, 10,108 alleles were absent in 1983–1993, 71,588 in 1994–2005, and 91,766 in 2006–2014. Correspondingly, the 1994–2005 sample lacked 20,002 unique alleles from the first and 24,070 from the second male founder, as well as 3518 alleles shared by both male founders. The sample from 2006–2014 lacked 34,189 and 28,735 unique alleles of the respective male founder, and lacked 4942 alleles shared by them. Per-chromosome results of absent alleles are provided in Supplemental Tables S5 and S6.
Similar to the temporal analyses of haplotypes, we examined to what extent lost SNP alleles “reappeared” in later time periods. Only 1% of the female founder alleles absent in 1983–1993, and 3% of her alleles absent in 1994–2005, were present in later time periods (Supplemental Table S7). Correspondingly, 3% and 29%, respectively, of the first and second male founder alleles absent in 1994–2005 were detected in 2006–2014. The accumulation of lost alleles over time is shown in Figure 3B. In general, the number of lost alleles agreed well with the number of the lost 1-Mb haplotypes (Fig. 3A).
Genomic distribution of lost haplotypes and alleles
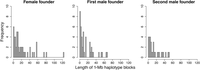
In an isolated population founded by just a few individuals, large segments from founder chromosomes can get lost from the population in the first generations of inbreeding before recombination has generated increasingly shorter haplotype blocks. The genomic distribution of absent 1-Mb founder haplotypes was in accordance with this prediction. The majority of lost 1-Mb haplotypes were concentrated into larger segments (Fig. 4), typically 0–3 per chromosome (Fig. 5). The size of these lost blocks varied from a single window to a whole chromosome (e.g., Chromosomes 1, 7, 17, and 25), with 2–5 Mb being the most common size. For several windows in the genomes of the female founder as well as the first male founder, both homologous haplotypes became lost.
Length distribution of lost 1-Mb haplotype blocks (x-axes represent the number of adjacent 1-Mb haplotypes in a block).
Chromosomal distribution of lost founder 1-Mb haplotypes. The majority of lost 1-Mb windows are clustered in larger segments. Haplotype clusters absent in the 1994–2005 subsample are in blue, and those absent in 2006–2014 are in black. Clusters absent in both 1994–2005 and 2006–2014 are in dashed blue. Haplotype clusters of the female founder that were absent already in 1983–1993 are outlined in red.
We intersected the location of lost 1-Mb haplotypes with the coordinates of lost SNPs. The vast majority (99%) of lost alleles from the founder female were clustered within lost haplotypes, providing an overall validation of the results from phasing (Supplemental Fig. S5). Similarly, 97% and 94% of lost alleles unique to the first and second male founder were concordant with coordinates of lost haplotypes. Approximately half of the SNP alleles that appeared only after 1993 were clustered within the two genomic regions on Chromosomes 19 and 20 that coincided with the 1-Mb haplotype blocks of the second male founder that were also observed only after 1993 (Supplemental Fig. S6).
The intersection of lost alleles and lost 1-Mb haplotypes was used to define more precise boundaries of lost chromosomal segments. This revealed that 3%, 19%, and 24% of the female founder's diploid genome was absent in the time periods 1983–1993, 1994–2005, and 2006–2014, respectively (Table 2). The corresponding proportions for the male founders were 10% and 8% in 1994–2005, followed by 15% and 10% in 2006–2014. Expressed as the amount of DNA, 1.096 Gb of the founder female's diploid genome had become lost by the third time period. Similarly, 681 and 468 Mb of the two male founders’ genomes became lost. The number of segregating alleles within these lost genomic regions was approximately 92,000, 39,000, and 34,000, respectively, which gives an indication of the significant genetic erosion caused by inbreeding and drift in this population.
Summarized parameters of absent chromosomal segments in the three temporal subsamples of Scandinavian wolves
Discussion
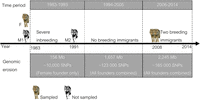
By analyzing 76 whole genomes from temporal subsamples of Scandinavian wolves, we illustrate the possibility of direct quantification of genomic erosion in a highly inbred natural population (Fig. 6). Our data reveal considerable loss of large haplotype segments, sometimes spanning entire chromosomes, and directly highlight genomic regions of low haplotype diversity.
Schematic summary illustrating population history and cumulative genomic erosion of the Scandinavian wolf population. The amount of DNA and number of SNPs lost in each time period are shown. (F) female founder, (M1) first male founder, (M2) second male founder. Breeding immigrants from 2008 and their offspring have been sampled but were not included in this study.
The contemporary Scandinavian wolf population has reduced levels of genetic diversity as a result of two processes. First, the genetic input at establishment in the 1980s and early 1990s was highly limited. Indeed, with just three founders, only a fraction of the genetic diversity of the source population in Finland, and possibly Russia, became represented in the Scandinavian population (Sundqvist et al. 2001; Vilà et al. 2003). Second, severe inbreeding and genetic drift further reduced the already limited diversity provided by the founders (no additional immigrant wolves reproduced within the Scandinavian population until 2008); only recently has there been some gene flow from the source population (Åkesson et al. 2016). We analyzed the effects of both inbreeding and drift.
Studies assessing and modeling inbreeding effects and loss of genetic diversity based on pedigree data usually assume that founder individuals are unrelated and outbred (Lacy 1989; Grueber and Jamieson 2008; Jansson and Laikre 2014; Bruford 2015). In the Scandinavian wolf population, this assumption would mean that six unique haplotypes have entered the population. However, in this study we show that this was not the case for most (75.6%) of the autosomal genome. One reason was that the three founders were not fully heterozygous, with 4%, 23%, and 23% of 1-Mb haplotypes being in homozygous state, respectively (but see below). There were extensive tracts of runs of homozygosity in each of these individuals. Another reason was that the three founders showed significant haplotype sharing. On average, there were 4.8 unique haplotypes per autosomal 1-Mb window, clearly violating the assumption of maximum founding diversity, which in this case would be six haplotypes.
Considering the amount and length of shared haplotypes, as well as the extent of homozygosity, it is likely that all three founders shared a common ancestor in a recent past. This is in accordance with the recent finding that Scandinavian wolves are on average more inbred than expected from pedigree-based relationships (Kardos et al. 2018). A similar situation has been suggested in the neighboring Finnish wolf population (based on allele frequency data) (Granroth-Wilding et al. 2017). Finland represents the edge of a large and probably continuous Russian–Finnish wolf population distributed across northern Eurasia (Linnell et al. 2008; Stronen et al. 2013; Bragina et al. 2015). Similar to the Scandinavian population, the Finnish wolf population decreased significantly in number during the 20th century, with several distinct bottlenecks (Pulliainen 1980; Ermala 2003), as has also been the case for wolf populations in continental Europe (Hindrikson et al. 2017; Dufresnes et al. 2018). This led to reduced levels of genetic diversity in the Finnish population (Jansson et al. 2012, 2014) and at least occasional cases of inbreeding (Granroth-Wilding et al. 2017). The fact that several immigrants show non-zero inbreeding coefficients estimated from runs of homozygosity (FROH) is also consistent with this (Kardos et al. 2018). As somewhat of an extreme case, two recent immigrants first reproducing in 2013 had FROH = 0.10 and 0.15, respectively, and their common offspring had FROH = 0.24 and 0.26, indicating incestuous mating between inbred individuals (Kardos et al. 2018).
When it comes to the roles of both genetic drift and inbreeding in the population, 10%–24% of the three founder genomes had become lost by the time of the most recent time period analyzed (2006–2014). In the case of the female founder (24%), this corresponds to a loss of more than 1 Gb of DNA that existed in the founding population. Approximately 92,000 SNP alleles unique to these lost regions of her genome disappeared from the population, and so did at least 73,000 alleles from the genomes of the male founders. The loss of some of these alleles may have consequences both for long-term survival and for counteracting short-term inbreeding depression.
The size of the Scandinavian wolf population grew considerably during 1983–2014. Until the third founder arrived to the population in 1991, there was at most one pack reproducing per year. Because the first two founders died early (the female founder in 1985), there were two generations of incestuous full-sibs and parent-offspring breeding as revealed from the pedigree (Åkesson et al. 2016). The arrival of a third founder resulted in an immediate genetic rescue effect and an increase in population size (Vilà et al. 2003). During the 1980s there were never more than 10 individuals, but the population increased to about 150 individuals in 2005 and about 460 individuals in 2014 (Anon 2015; Åkesson et al. 2016). This means that the strength of genetic drift during the first time period of our study (1983–1993) should have been most pronounced for the female and the first male founder lineages, and the number of haplotypes recorded as lost therefore highest in the following time period (1994–2005). This was exactly what we observed. The number of lost haplotypes in 1994–2005 exceeded that in both 1983–1993 (data only available for the female founder) and 2006–2014 (female founder and first male founder).
The finding of significant variation in founder haplotype diversity across the genome illustrates the importance of performing whole-genome analysis. More limited sampling of markers or genomic regions may have led to a biased picture of both the extent of starting levels of diversity and the subsequent loss of genetic diversity. Genomic regions of low founder diversity should be at highest risk for fixation and potentially inbreeding depression. Incidentally, the two genomic segments with the lowest founder haplotype diversity (two unique haplotypes) were a 5-Mb genomic region on Chromosome 12 harboring the major histocompatibility complex (MHC) loci (cf. Seddon and Ellegren 2002; 2004), and a 5-Mb genomic region on Chromosome 33 harboring olfactory receptor (OR) genes, among others. High levels of polymorphism at MHC and OR genes are thought to be important for long-term survival of populations (Sommer 2005; Tacher et al. 2005; Robin et al. 2009; Niskanen et al. 2013). Although the frequency of the two MHC haplotypes was similar in all three temporal groups (see Seddon and Ellegren 2004), drift led one of the two OR haplotypes to segregate at very low frequency in the 2006–2014 sample (Supplemental Fig. S4).
Chromosome-level statistical phasing was essential in this study. It allowed the assignment of individual marker alleles into six parental haplotypes (including those of both male founders that were not sequenced), as well as detection of loss of genomic segments identical to other founder haplotypes. Even though whole-genome statistical phasing is challenging and might be error-prone (Andrés et al. 2007), it is cost-effective compared to read-based phasing for population scale analysis. The very high concordance between the genomic location of lost SNP alleles and the location of lost 1-Mb window haplotypes in each temporal subsample provides overall support to the robustness of our results. However, the extent of homozygosity in the nonsampled male founders may have been somewhat overestimated. Windows in which one and the same haplotype was consistently transmitted to their offspring were assigned as homozygous. However, it cannot be fully excluded that such windows were in fact heterozygous and that the other haplotype either directly got lost from the population or remained undetected in our sample. It is difficult to quantify this possible source of bias.
Another methodological aspect is that a haplotype recorded as absent in a particular time period could still be present in the population but segregating at low frequency and elude detection in the investigated sample. This would lead to an overestimation of the loss of diversity. However, for the female founder (1.8%–3.1%) and the first male founder (5.8%), the proportion of 1-Mb haplotypes noted as absent in one time period but appearing in a later was low. In the case of the second male, on the other hand, 32.5% of his inferred haplotypes not detected in 1994–2005 were seen in the 2006–2014 sample. This can probably be explained by the sampling strategy when choosing individuals for sequencing. Most of the “reappearing” haplotypes in 2006–2014 were clustered in five larger haplotype blocks (Fig. 5, genomic segments in blue) that were found in one or two closely related individuals whose parents from 1994–2005 were not sequenced.
With the extensive loss of genetic diversity documented here and without gene flow from neighboring populations, Scandinavian wolves would clearly be in genetic peril. Meeting conservation goals such as retaining at least 95% of heterozygosity over 100 yr (Allendorf and Ryman 2002) would obviously have been impossible. Moreover, the presence of strongly deleterious mutations would be associated with a high risk of extinction (see Kyriazis et al. 2021). Immigrant wolves have regularly been recorded in the Scandinavian wolf population since its reestablishment in the 1980s (Seddon et al. 2006; Åkesson et al. 2016). Most of these immigrants have failed to establish genetic contact with the local population because their appearance in reindeer herding areas have legalized protective hunt. Starting in 2008, however, a handful of immigrants have become integrated with the local population (as indicated above, descendants of these recent immigrants were not included in this study) (Åkesson et al. 2016). Still, inbreeding levels continue to be high (Åkesson et al. 2016) and signs of inbreeding depression have been recorded. This includes reduced litter size and/or juvenile survival (e.g., see Figs. 3 and 5; Wabakken et al. 2001; Liberg et al. 2005), age at first reproduction (Wikenros et al. 2021), as well as congenital anomalies (Räikkönen et al. 2006; 2013). It will be important in the future to monitor the spread of incoming haplotypes as well as to follow the survival of founder haplotypes.
In conclusion, this study presents a novel genomic approach to quantify the loss of genetic diversity across time in an endangered mammal population. We show the strength of phased data for resolving the ancestry of genomic variants and for pinpointing specific genomic regions of low diversity. Empirical insight into limited founder diversity and extensive haplotype loss emphasize the importance of gene flow to counteract genomic erosion of small populations.
Methods
Samples
The study comprised 76 Scandinavian wolves sampled between 1984 and 2015. Illumina short-read, whole-genome sequence data from 73 individuals were obtained from Kardos et al. (2018) available at the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) under accession number PRJEB20635, and three additional individuals were resequenced as described in Kardos et al. (2018). Briefly, DNA was prepared from blood or muscle tissue and paired-end libraries constructed for 150-bp sequencing on an Illumina HiSeq X instrument.
The material included the female founder of the population and 75 individuals born in Scandinavia (1983–2014) whose ancestry trace back solely to the three founder wolves (Supplemental Table S8). The two founder males were never sampled. Compared to the larger set of wolves analyzed by Kardos et al. (2018), recently reproducing immigrants (including two males in 2008, and one male and one female in 2013) and their descendants, as well as immigrants that never reproduced in Scandinavia (as identified by Åkesson et al. 2016) were not included in this study. For the purpose of statistical phasing, we also included whole-genome sequence data from 98 Finnish wolves (Smeds et al. 2019, 2021) available at ENA under accession numbers PRJEB28342 and PRJEB39198.
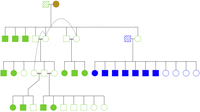
In temporal analyses of loss of genetic diversity, we divided the data set into three time periods: wolves born in 1983–1993 (n = 19 individuals), 1994–2005 (n = 28), or 2006–2014 (n = 28). The individuals from 1983 to 1993 consisted of 12 F1–F3 generation offspring of the first male founder and seven F1 offspring of the second male founder. This is before descendants from the two male founder lineages established breeding pairs with each other, making it a suitable group for tracing male founder alleles (Fig. 7). For the remaining time period up until 2014, wolves were split in two temporal subgroups of equal sample size (i.e., 1994–2005 and 2006–2014). The birth year of individual wolves was estimated based on aging using one of three methods: (a) tooth root sectioning and counting cementum annulation (Landon et al. 1998; Gipson et al. 2000) at Matson's Laboratory (https://matsonslab.com/the-science/cementum-aging/); (b) aging based on years of parental reproduction (Wikenros et al. 2021); or (c) morphological determination of juveniles (<1 yr) at the postmortem using macroscopic analyses of dentition and identification of bone growth plates by radiography.
Pedigree of the first offspring generations, born 1983–1993, of the Scandinavian wolf population. The female founder is shown with a filled brown circle, the first male founder with a green stripe pattern square, and the second male founder with blue stripe pattern square. Sequenced offspring of the first male founder are shown as filled green symbols, and of the second male founder as filled blue symbols; not sequenced offspring are left white. Dashed lines link three different mating pairs of the same female, and double lines represent breeding pairs of close relatives. The figure was prepared using R version 3.3.3 (https://cran.r-project.org/bin/windows/base/old/3.3.3/) (R Core Team 2017) and kinship2 v.1.6.4 (https://cran.r-project.org/web/packages/kinship2/index.html).
Variant calling and filtering
Alignment of whole-genome sequence data and joint variant calling of Scandinavian and Finnish wolves was performed in accordance with Kardos et al. (2018) and Smeds et al. (2021) using the CanFam3.1 genome assembly (Lindblad-Toh et al. 2005). The obtained data set of genomic variants was further subjected to stringent filtering. First, we combined known coordinates of transposable elements and windows of highly repetitive sequence in the CanFam3.1 genome assembly (Lindblad-Toh et al. 2005) and excluded these regions from the analysis. Genomic coordinates obtained by RepeatMasker (Smit et al. 1996-2010) and WindowMasker (Morgulis et al. 2006) were downloaded from the UCSC Table browser (Karolchik et al. 2004). Second, to eliminate potential ambiguities in SNP calling caused by segmental duplications in the genome assembly, each chromosome was self-aligned with LASTZ v.1.04 (Harris 2007), and variants located within self-aligned regions were excluded. Third, remaining markers were filtered with the following criteria “‐‐remove-indels ‐‐mac 1 ‐‐min-alleles 2 ‐‐max-alleles 2 ‐‐minGQ 30 ‐‐minQ 300 ‐‐maxDP 80” using VCFtools 0.1.15 (Danecek et al. 2011). Fourth, we discarded SNP markers coinciding with SNP-dense genomic regions (>7 SNPs/kb). The remaining data set after filtering consisted of 3,900,583 SNP markers. Individual chromosomes of the reference genome were indexed by SAMtools 1.8. (Li et al. 2009).
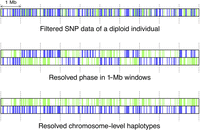
Two-step statistical phasing
For statistical phasing we retained a subset of SNP markers that had <25% missing genotypes and were at least 20 kb apart from each other (107,576 SNPs). This was done to reduce computational load and the occurrence of imputation and genotyping errors. Filtered genomic variants of Scandinavian and Finnish wolves were jointly phased by PHASE 2.1.1. (Stephens et al. 2001), a statistical haplotype reconstruction tool in two separate steps based on observed population data (Fig. 8). In the first phasing step each chromosome of the CanFam3.1 reference assembly was split into nonoverlapping 1-Mb windows by BEDTools version 2.27.1 (Quinlan and Hall 2010). The chosen window size was a compromise between the precision and accuracy of phasing, as window sizes smaller than 1-Mb resulted in an increased number of switch errors during the second phasing step. The CanFam3.1 reference assembly has sequence assigned to 38 autosomes and the X Chromosome and comprise a total length of 2,327,633,984 bp. We obtained data from 2344 1-Mb windows, including the last window of each chromosome smaller than 1-Mb if phasing was informative. Each window was then independently phased with biallelic SNP marker settings. In the second step each individually phased 1-Mb region was considered as one multiallelic locus where alleles correspond to phased 1-Mb haplotypes. Thus, statistical phasing of chromosome-level haplotypes was subsequently done with multiallelic settings. A custom Perl script was used to convert the PHASE output from the first phasing step to input for the second step (Supplemental Code).
A schematic overview of two-step phasing. Green and blue lines represent nonreference SNP alleles from the two haplotypes of a single diploid individual.
To infer chromosome-level haplotypes of the unsampled male founders, we first identified haplotypes from the female founder in her sequenced F1–F3 offspring from 1983 to 1993. Remaining haplotypes different from those found in the female founder were assigned to the respective male founder based on pedigree information (Åkesson et al. 2016). As the second male founder bred with an F1 female of the first male founder, haplotypes of the first male founder were in some cases derived from F1 offspring of the second male founder (Chromosomes 7 and 19). At two occasions (Chromosomes 19 and 20), haplotypes of the second male founder were derived from his F2 offspring in 1994–2005. All 1-Mb haplotypes in offspring from 1983 to 1993 identical to those of the female founder embedded within chromosome-level haplotypes harboring 1-Mb haplotypes specific to the first male founder were considered as shared between him and the female founder. Similarly, all 1-Mb haplotypes identical to those of the female founder, or the first male founder, embedded within chromosome-level haplotypes of the second male founder were considered to be shared between him and the female or the first male founder. After obtaining all six founder haplotypes, we assigned the founder origin of 1-Mb haplotypes for all individuals born in 1994–2014.
Manual curation of the phased data set
Given three founders, we expect to observe up to six unique autosomal haplotypes entering the population and up to four haplotypes for the X Chromosome. Additional haplotypes detected in subsequent generations must be either founder haplotypes not observed in 1983–1993 or have been generated by recombination. However, phasing errors will also result in what appears as new haplotypes. The chromosome-level phased data set was manually curated to discriminate between new haplotypes arising by recombination from those resulting from phasing or SNP genotyping errors.
As a general guideline for curation, we analyzed if flanking windows of each side of a new haplotype came from different founder haplotypes, which would be consistent with recombination. If a new haplotype was embedded within a single haplotype, it was considered a potential phasing error. We further investigated such cases by inspecting the marker phasing probabilities within the particular window and zygosity of the neighboring markers and tested whether removal of poorly phased markers would improve the phase concordance across the neighboring windows. The main contributors to phasing errors were missing data resulting in falsely imputed haplotypes and SNP genotyping errors, for example, caused by segmental duplications in the genome of the resequenced individual or the alternative allele not being sequenced. Nearby double recombination and gene conversion events would also mimic the pattern of new haplotypes but were neglected in this study. In cases when phase of a certain window remained unresolved after manual correction, this window was removed from further analysis (11 out of 2344 windows). Recombinant windows retained their unique haplotype IDs, whereas erroneously phased windows were manually assigned with a founder haplotype ID.
Presence/absence analysis of SNP alleles
We sought to follow the survival of individual alleles over time, and for the sequenced female founder this was straightforward. Because the male founders were not sequenced, their respective allele contribution was obtained by recording alleles that were present in their offspring from the 1983–1993 sample, as described above, but not carried by the female founder. With this approach we only recorded alleles that are unique to male founders and were not able to identify those alleles common between the female founder and one or both of the males. Also, alleles of the first male founder that occurred only in offspring of the second male founder would be assigned to the second male founder. To reduce the risk of missing alleles unique to the male founders, we discarded markers in which at least one individual from 1983 to 1993 had a missing genotype. Here, in comparison to our initial joint variant calling step of Scandinavian and Finnish wolves, we removed all sites that were fixed in the Scandinavian population. Thus, the filtered data set included only 1,479,905 out of 3.9 million SNPs called at the initial variant calling step. The presence/absence of genotyped alleles was scored with a set of in-house scripts (Supplemental Code).
Data access
Raw sequence data generated in this study have been submitted to the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) under accession number PRJEB44869.
Competing interest statement
The authors declare no competing interests.
Acknowledgments
We thank Linnéa Smeds for help throughout the project. This work was supported by grants from the Knut and Alice Wallenberg Foundation and the Swedish Research Council to H.E.
Author contributions: H.E. conceived the study; A.V. and H.E. designed the study; A.V. performed the research; Ø.F., M.Å., C.W., H.S., and P.W. provided samples and information about them; A.V. wrote the paper together with H.E.; and all authors reviewed and edited the manuscript.
Footnotes
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276070.121.
-
Freely available online through the Genome Research Open Access option.
- Received August 3, 2021.
- Accepted December 30, 2021.
This article, published in Genome Research, is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.