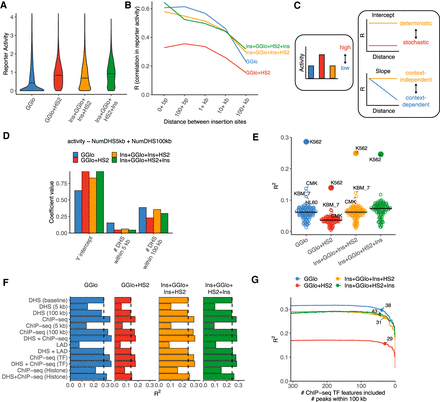
Quantitative assessment of genomic context effects on reporter activity. (A) Distribution of activity by reporter class. Average activity was computed across all fully mappable 10-kb bins with at least one insertion. Horizontal bars represent medians. (B) Correlation in activity for nearby insertions by reporter class merged across all experiments. For each insertion, 50 nearby insertions were sampled with replacement from within 500 kb. Correlation was computed across pairs of insertions in each distance bin. Bins with fewer than 100 data points were omitted. (C) Model of genomic context effects on reporter activity and correlation profiles. Increased correlation across all length scales reflects deterministic versus stochastic activity. Constant correlation across all length scales reflects context-independent activity, whereas a reduction of correlation with distance represents genomic context dependency. (D) Linear regression coefficients for model partitioning reporter activity into close and long-range genomic context represented by count of DHSs within 5 and 100 kb, respectively. (E) Predictive performance of reporter activity using DHS data from other cell types. Bar indicates median. (F) Predictive performance of models incorporating ENCODE histone and TF ChIP-seq data, and lamin-associated domains (LADs) (Leemans et al. 2019). Model including the number of DHSs within 5 and 100 kb as features is used as baseline for comparison (dashed line). (G) Analysis of TF ChIP-seq feature importance under iterative removal of feature with smallest effect size. Inflection points are labeled with the number of ChIP-seq features.











