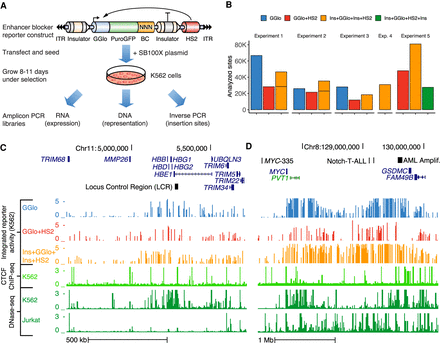
Site-specific profiling of enhancer blocker activity. (A) Barcoded reporters were randomly integrated using SB100X transposase. Site-specific reporter activity was read out in multiplex using sequencing to map insertion sites (inverse PCR libraries), barcode representation (DNA libraries), and expression (RNA libraries). (GGlo) HBG1 promoter; (BC) unique barcode; (HS2) beta-globin hypersensitive site 2 enhancer; (ITR) Sleeping Beauty inverted terminal repeats. (B) Counts of analyzed sites in thousands for five experiments including promoter-only (GGlo), promoter and HS2 enhancer (GGlo + HS2), with CTCF site interposed between GGlo and HS2 (Ins + GGlo + Ins + HS2), or with CTCF sites fully flanking the reporter and enhancer (Ins + GGlo + HS2 + Ins). (C,D) Analysis of enhancer blocker functionality at the HBB (C) and MYC (D) loci. The top three tracks show reporter activity. Data shown merged from replicate experiments. The bottom three tracks show CTCF ChIP-seq data for K562 erythroleukemia cells, and DNase-seq data for K562 and Jurkat T-cell leukemia cells. Regions highlighted in D include the MYC-335 enhancer region coinciding with a genetic association for colorectal cancer (Sur et al. 2012), the Notch-T-ALL (Acute Lymphocytic Leukemia) enhancer cluster (Herranz et al. 2014), and the AML (acute myeloid leukemia) amplified region (Radtke et al. 2009).











