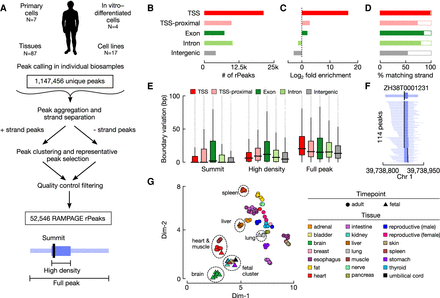
Curating a collection of representative RAMPAGE peaks (rPeaks) across 115 biosamples. (A) Workflow for curating RAMPAGE rPeaks. First, we called peaks in individual RAMPAGE data sets across 115 cell types and tissues. We then pooled these peaks (N = 1,147,456) and separated them by genomic strand. We clustered overlapping peaks on the same strand, selected the peak with the highest RAMPAGE signal (i.e., the rPeak) to represent each cluster, removed all the peaks overlapping the rPeak from the pool, and performed clustering on the remaining peaks. We repeated this process iteratively until all peaks were accounted for by rPeaks. We performed additional filtering using RNA-seq data, removing peaks that had a higher RNA-seq signal than RAMPAGE signal, finally arriving at 52,546 rPeaks. (B) Bar plots showing the number of RAMPAGE rPeaks stratified into distinct sets by genome context: overlapping GENCODE V31 TSSs (red), proximal (±500 bp) to TSSs (pink), overlapping exons (dark green), overlapping introns (light green), and intergenic (gray). (C) Bar plots showing the fold enrichment for the number of genomic positions covered by rPeaks over the footprints of the genomic contexts in B. (D) Bar plots showing the percentage of rPeaks in each genomic context as in B that are on the same strand as their overlapping TSS, gene (exon and intron), or nearest gene (TSS-proximal and intergenic). (E) Box plots displaying the variation in the positions of rPeak summits (left), high-density region boundaries (middle), and full peak boundaries (right), stratified by the genomic contexts as in B. (F) An example TSS-overlapping rPeak ZH38T000123 from K562 cells and the RAMPAGE peaks it represents in 113 other biosamples. For each peak, the full width is denoted in light blue, high-density regions in blue, and summit in black. (G) Scatterplot displaying a two-dimensional Uniform Manifold Approximation and Projection (UMAP) embedding of 87 tissue samples using RAMPAGE signal across all rPeaks as input features. Circles denote adult tissues, and triangles denote fetal tissues. Markers are colored by tissue of origin as defined in the legend.











