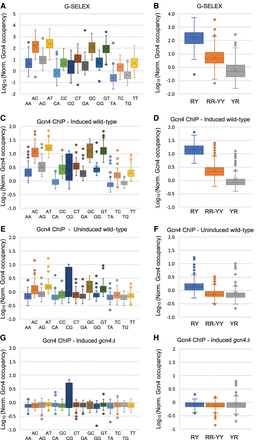
The high-affinity Gcn4 binding site is RTGACTCAY: Comparison of AP-1 site binding as a function of flanking base pairs. All 1078 AP-1 sites (TGACTCA) were subdivided and named according to their flanking base pairs (e.g., “AT” indicates the motif ATGACTCAT). Data were normalized to the genomic average (=1) and decompressed using a log scale. (A) G-SELEX data for each of the 16 possible types of AP-1 site. (B) G-SELEX data for the three possible motif types with R = A or G and Y = C or T. (C,D) Gcn4 ChIP-seq data for induced wild-type cells. (E,F) Gcn4 ChIP-seq data for uninduced wild-type cells. (G,H) Gcn4 ChIP-seq data for induced gcn4Δ cells.











