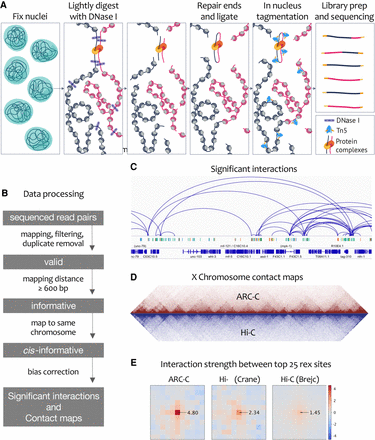
ARC-C method and comparison with Hi-C. (A) ARC-C cartoon. (B) Data processing steps. (C) Integrative Genomics Viewer (IGV) (Robinson et al. 2011) screen shot showing significant interactions in a 275-kb window of Chr III (4,075,000–4,300,822). Regulatory elements (protein-coding promoters, red; unassigned promoters, yellow; enhancers, green; unknown activity, blue) (Jänes et al. 2018) and genes are displayed below. (D) Comparison of ARC-C and Hi-C X Chromosome contact maps. Hi-C data are from Crane et al. (2015). (E) Aggregate contact analysis plots (Rao et al. 2014) showing signal between top 25 rex sites (Crane et al. 2015) at 10-kb resolution and a distance range of 100 kb to 4 Mb for ARC-C (this study) and two C. elegans Hi-C maps (Crane et al. 2015; Brejc et al. 2017). Arrows indicate the linear enrichment of rex–rex interactions (at the central 10-kb square) relative to interactions between other regions. rex–rex interaction strength was statistically significant (P < 0.001; permutation test, see Methods) in all three maps.











