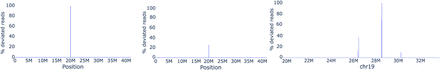
Distance concordance test applied to simulated and real read-sets. (Left) Mapping reads from the data set Cen9Sim reveals an artificial deletion at location 20 Mbp. Concordance(aA, bA) = 1 and TopDiff(aA, bA) = 10,003 for solid k-mer aA at position 19,999,691 and bA at position 20,000,001, revealing the approximate location of the deletion of size 10 kbp. (Middle) Mapping reads from the data set Cen9Sim-Het reveals an artificial Het site at position 20 Mbp with Concordance(aA, bA) = 0.33 for the same solid k-mers aA and bA and TopDiff(aA, bA) = 10,005 pointing at insertion in Cen9del10 of size 10 kbp existing in one of the artificial haplotypes. (Right) Mapping HiFi reads reveals several Het variants and a likely assembly error (with Concordance = 1 at 28.5 Mbp) in the centromere of Chromosome 19 (for description of all data sets, see Supplemental Table 1; Supplemental Note 4, “Extended benchmarking of long-read mapping tools”).











