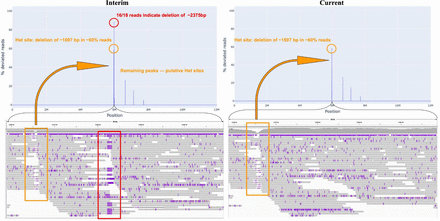
VerityMap detects a deletion in CHM13-Cen10-interim that is absent from the publicly released CHM13 assembly. The top panel shows the distance concordance test applied to CHM13-Cen10-interim (left) and Cen10 in CHM13 assembly (right). The bottom panel shows read alignments ∼6 Mbp in two versions of the assembly. (Bottom left) VerityMap reveals a deletion of length ∼2.4 kbp in CHM13-Cen10-interim. The remaining peaks correspond to the Het sites with various multiplicities in the assembly. In particular, one heterozygous deletion of ∼1.7 kbp that is supported by ∼60% of reads is only 20 kbp upstream of the misassembly site. (Bottom right) The misassembly is corrected in the current version of Cen10 assembly, whereas the putative Het sites remain.











