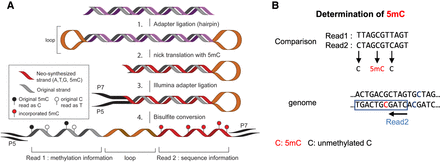
Overview of Methyl-SNP-seq. (A) Experimental workflow of Methyl-SNP-seq: (1) The genomic DNA is fragmented to 300–400 bp fragments. (2) Hairpin adapters are ligated at both ends of the fragmented DNA, forming a dumbbell-shaped DNA. Next, nicks at both opposite ends of the adapters are introduced and using nick translation, a copy of the original strand is synthesized, replacing CTP as a source of nucleotide with m5CTP instead. This nick translation step breaks the dumbbell-shaped DNA somewhere within the fragment. (3) Methylated Illumina Y-shaped adapters are ligated. (4) Bisulfite conversion opens the DNA structure revealing a single-stranded DNA molecule that can be amplified using the Illumina adapters. Sequencing requires paired-end reads to obtain both the methylation and the genomic sequence information. For more details on the experimental procedure, see Supplemental Figure 1A and Supplemental Protocol. (B) Deconvolution procedure: for more details on the bioinformatics analysis, see Supplemental Figure 1B.











