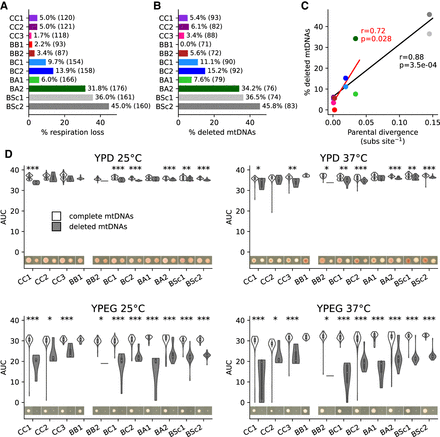
Frequent deletions in mtDNAs lead to the loss of respiratory function and occur preferentially in crosses with higher parental divergence. (A) Percentage of all MA lines with loss of respiration. Counts of lines (at initial and final time points combined) are shown. (B) Percentage of sequenced MA lines with mtDNA deletions. Counts of lines (initial and final time points combined) are shown. (C) Pearson's correlation between percentage of mtDNAs with deletions and parental divergence for each cross (substitutions per site, based on nuclear genome-wide variants). The correlation shown in red excludes BSc crosses. (D) Area under growth curves (AUC) for lines with complete and deleted mtDNAs in the four tested conditions. FDR-corrected P-values for Mann–Whitney U tests between complete and deleted mtDNAs: (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001. Images show colonies of representative lines (i.e., the closest to the median) after 4 d.











