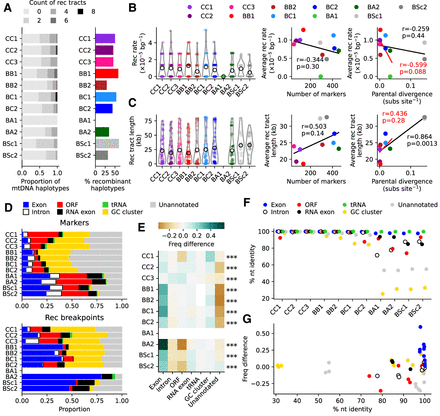
mtDNA protein-coding exons are enriched in recombination junctions. (A, left) Counts of recombination tracts per mtDNA haplotype. (Right) Percentages of recombinant haplotypes. (B, left) Recombination rate distributions for individual lines. Hollow circles show average recombination rates. (Right) Pearson's correlation between the average recombination rate and the number of markers or parental evolutionary divergence (substitutions per site, based on nuclear genome-wide variants). The correlation in red excludes BSc crosses. (C, left) Length distributions for individual recombination tracts. Hollow circles show average recombination tract lengths. (Right) Pearson's correlation between the average recombination tract length and the number of markers or parental evolutionary divergence (substitutions per site, based on nuclear genome-wide variants). The correlation in red excludes BSc crosses. (D) Distributions of annotation feature types for marker variants (top) and recombination breakpoints (bottom). (E) Overrepresentation of each annotation feature type in recombination junctions, calculated as the difference in frequency between breakpoints and markers for each feature type. FDR-corrected P-values of chi-squared tests for frequency deviations within each cross are shown: (***) P ≤ 0.001. (F) Percentage of nucleotide identity between parental mtDNAs for each annotation feature type. (G) Overrepresentation of each annotation feature type in recombination junctions against the percentage of nucleotide identity. Values for each cross are represented as separate points.











