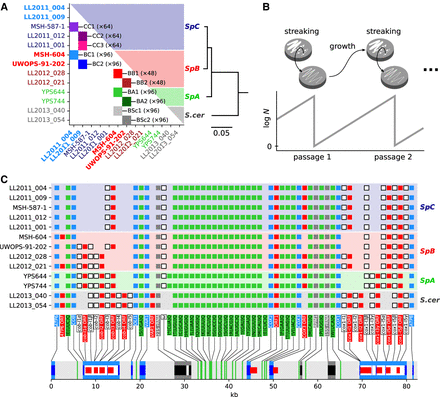
Design of the MA experiment on Saccharomyces hybrids and mtDNA contents of the parental strains. (A) Design of the MA crosses. Colored squares indicate crosses between the corresponding parental strains on both axes. Strains in bold are shared by multiple crosses. Eight hundred sixty-four independent MA lines were subdivided into 11 crosses spanning intra-lineage (S. paradoxus SpC×SpC [CC]; S. paradoxus SpB×SpB [BB]), intraspecific (S. paradoxus SpB×SpC [BC]; S. paradoxus SpB×SpA [BA]), and inter-specific (S. paradoxus SpB× S. cerevisiae[BSc]) parental divergence. Each cross was replicated to initiate 48 to 96 independent evolution lines, all arising from distinct mating events. The phylogenetic tree summarizes the evolutionary divergence between parental strains (substitutions per site, based on nuclear genome-wide variants). (B) Microbial MA experiments minimize the efficiency of natural selection. Periodic extreme bottlenecks are achieved through streaking for single colonies on solid medium, which amplifies genetic drift. (C) Annotation summary of high-quality reference mtDNA assemblies for each parental strain. Colored squares denote the presence of each feature in the corresponding genome. Light blue indicates protein-coding genes; dark blue, protein-coding exons; white, introns; gray, RNA-coding genes; black, RNA-coding exons; red, intronic or free-standing ORFs; and green, tRNAs.











