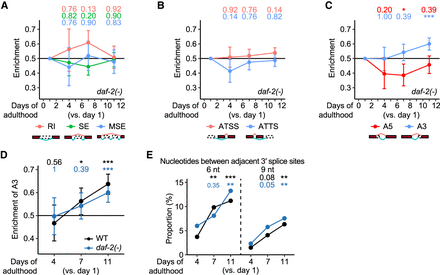
daf-2 mutations slow age-dependent increases in the usage of distal 3′ splice sites in transcript isoforms. (A–C) Enrichment of different RNA-processing events in isoform fractions that were up-regulated among isoform fractions changed during aging in daf-2(e1370) [daf-2(-)] animals. RNA-processing events include retained introns (RI), skipped exons (SE), multiple skipped exons (MSE) (A), alternative transcription start sites (ATSS), alternative transcription termination sites (ATTS) (B), alternative 5′ splice sites (A5), and alternative 3′ splice sites (A3) (C). (D) Enrichment of A3 in isoforms that were up-regulated among isoforms whose fractions changed during aging in wild-type (WT) and daf-2(-) animals. Adjusted P values are shown at the top of the data points, calculated by one-proportion Z-test and adjusted using false discovery rates; (*) adjusted P < 0.1, (**) adjusted P< 0.01, (***) adjusted P < 0.001. Note that results of WT are the same as those used in Figure 2E, and shown here for comparison with those of daf-2(-) animals. (E) Proportion of the proximal and distal 3′ splice sites separated by 6 nt and 9 nt in age-dependently up-regulated A3 isoforms in WT and daf-2(-) animals. P values are shown at the top of the data points, calculated by using two-tailed Fisher's exact test; (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. See Supplemental Figure S11 for detailed analysis results of A3 isoforms in daf-2(-) animals.











