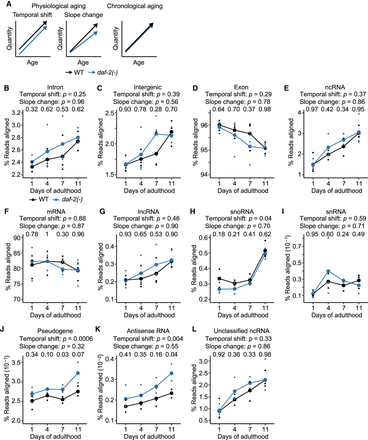
daf-2 mutations affect age-dependent changes in transcriptional fidelity. (A) Schematic of transcriptomic features that match physiological and chronological aging in wild-type (WT) and daf-2(e1370) [daf-2(-)] animals. The transcriptomic features associated with physiological aging were categorized as temporal shift that begins at early adulthood and slope change that alters aging rates throughout adulthood. (B–D) Overall expression levels of structural elements, including intron (B), intergenic (C), and exon regions (D) in WT and daf-2(-) animals at different ages. (E,F) Overall expression levels of functional elements, including noncoding RNA (ncRNA) (E) and protein-coding messenger RNA (mRNA) (F) in WT and daf-2(-) animals at indicated ages. (G–L) Age-dependent level changes in ncRNA, including long ncRNA (lncRNA) (G), small nucleolar RNA (snoRNA) (H), small nuclear RNA (snRNA) (I), pseudogene-coded RNA (pseudogene) (J), antisense RNA (K), and unclassified ncRNA (L) in WT and daf-2(-) animals. Overall snoRNA levels increased with age and shifted down in daf-2(-) animals compared with those in WT animals at day 1 of adulthood (H). The levels of pseudogene-coded RNAs (J) and antisense RNAs (K) increased with age and shifted up in daf-2(-) animals at day 1 of adulthood (Supplemental Fig. S5D,E). P value is shown at each day, calculated by two-tailed Welch's t-test between WT and daf-2(-) animals of the same chronological age. In each panel, two P values are shown for the effects of genotypes on transcript levels that correspond to “Temporal shift,” and those for interaction between genotypes and ages that correspond to “Slope change,” calculated by using two-way analysis of variance (ANOVA). Note that results from WT animals are the same as those used in Figure 1B–L; they are shown here for the comparison with daf-2(-) animals.











