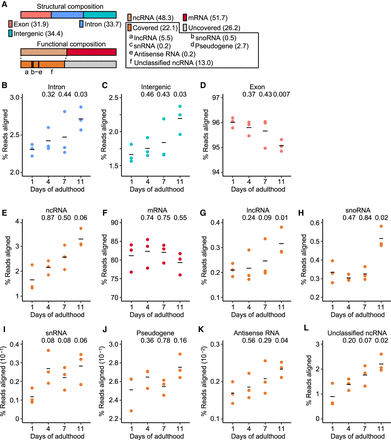
Reduced transcriptional fidelity is pervasive during aging in Caenorhabditis elegans. (A) Structural and functional compositions (%) of genomic elements in C. elegans. Structural composition includes exon, intron, and intergenic regions. Functional composition includes noncoding RNA (ncRNA) and protein-coding messenger RNA (mRNA). ncRNA is further divided into long ncRNA (lncRNA), small nucleolar RNA (snoRNA), small nuclear RNA (snRNA), pseudogene-coded RNA (pseudogene), antisense RNA, and unclassified ncRNA. Numbers in parentheses indicate relative proportions in the composition. “Covered” indicates ncRNAs that can generally be detected with RNA-seq, whereas “Uncovered” indicates ncRNAs that require specialized procedures for detection. (B–D) Overall expression levels of structural elements during aging: intron (B), intergenic (C), and exon regions (D). (E–L) Age-dependent changes in the expression levels of functional elements, including ncRNA (E), mRNA (F), lncRNA (G), snoRNA (H), snRNA (I), pseudogene-coded RNA (J), antisense RNA (K), and unclassified ncRNA (L). P value is shown on top of each panel. Two-tailed Welch's t-test relative to the data obtained with animals at day 1 of adulthood.











