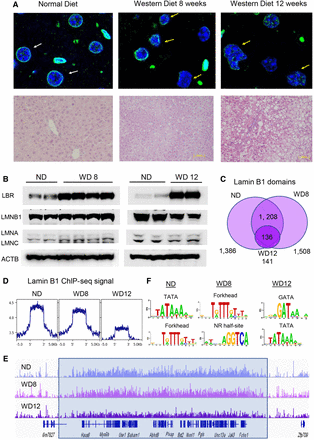
Nuclear lamina changes in diet-induced fatty liver. (A) Nuclear immunofluorescence (lamin B1; green) and DAPI nuclear staining of FFPE liver sections from mice on normal diet (ND; top left), WD8 (top middle), and WD12 (top right). Nuclei in mice on ND have a round shape (white arrows), whereas nuclei in mice on WD are irregular in shape and distorted (yellow arrows). Representative liver sections from ND and WD mice stained with H&E (bottom). Lipid accumulation is apparent on histological sections by the presence of lipid droplets in livers of mice on WD at 12 wk. (B) Western blot analysis of protein nuclear extracts from two control livers (ND) and four livers from mice on WD8 (left) and two control livers (ND) and two livers from mice on WD12 (right) with antibodies to LBR, LMNB1, LMNA/LMNC, and ACTB (loading control). Protein expression of LBR is increased in WD8, whereas that of LMNA, LMB1, and LMNC is not altered (left). Protein expression of LBR is increased and LMNB1 (lamin B1) decreased in WD12, whereas that of LMINA and LMNC is not changed (right). (C) Venn diagram showing the results of genome-wide location analysis for lamin B1 (ChIP-seq) in livers on ND, WD8, and WD12, identifying 1396 domains in ND, 1508 in WD8, and 141 in WD12 called bound in both young and old livers by SICER. The number of LADs is comparable in WD8 (1396 ND vs. 1508 WD8). The number of LADs is greatly reduced in WD12 (1396 ND vs. 141 WD12). (D) Lamin B1 ChIP-seq signal (reads per kilobase, per million mapped reads [RPKM]) calculated at LADs decreases with WD progression. (E) ChIP-seq track view in the Integrative Genome Viewer (IGV) of a representative LAD showing decreased lamin B1 ChIP-seq signal for WD at 12 wk. (F) Scanning motif of positional weight matrices in the JASPAR and TRANSFAC databases in LADs in ND (left), WD8 (middle), and WD12 (right). PscanChIP identified highly enriched consensus sites for A/T-rich sequences, including TATA box (P-value < 8.9 × 10−108) and the forkhead motif (P-value < 8.8 × 10−93) in ND, forkhead consensus (P-value < 4.9 × 10−106) and nuclear receptor half-site (P-value < 6.7 × 10−32) in WD8, and GATA consensus (P-value < 1.4 × 10−8) and A/T-rich TATA box motif (P-value < 1.8 × 10−5) in WD12.











