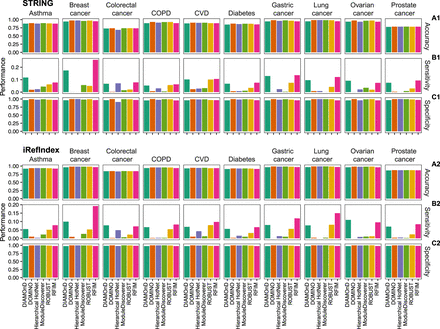
Disease association for genes in disease module detected from integrating the genome-wide association studies (GWAS) with two interactomes for each phenotype and method. Accuracy (A), Sensitivity (B), and Specificity (C) between the genes in the disease module and the gold standard data set of disease-gene associations from DisGeNET. We treated the disease-associated genes of each phenotype in DisGeNET as positive instances and the remaining genes as the negative instances. Similarly, the genes in the disease module were considered as positive instances and the remaining genes were considered as negative instances.











