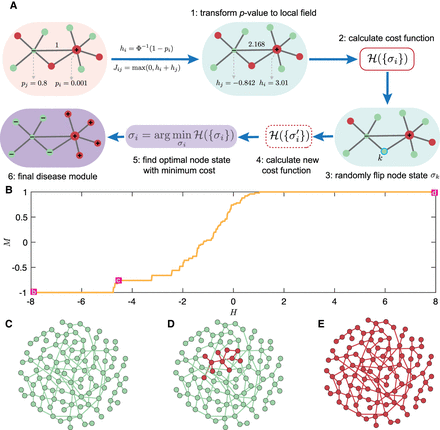
Disease module inference using statistical physics approach: a demonstration. (A) Schematic demonstration of the Random-field Ising model (RFIM) approach consisting of the following steps: (1) Given a graph
and gene-wise P-value associated with each node, we first transform P-value to node weight and assigned weight to each edge. (2) For any gene state {σi}, we calculated the cost function 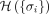 . (3) An optimization method was used to find the optimal gene state {σi} that minimizes the cost function. (4) All genes with state σi = 1 are those genes in the disease module. (B) The magnetization curve M(H) of the random network system. A random network with N = 100 nodes (genes). Each gene is assigned a P-value, randomly chosen from a uniform distribution U(0, 1). The node weights hi are then computed using the inverse normal CDF. The edge weights Jij = max(0, hi + hj) if two genes are connected in the network. Otherwise, Jij = 0. The magnetization
. (3) An optimization method was used to find the optimal gene state {σi} that minimizes the cost function. (4) All genes with state σi = 1 are those genes in the disease module. (B) The magnetization curve M(H) of the random network system. A random network with N = 100 nodes (genes). Each gene is assigned a P-value, randomly chosen from a uniform distribution U(0, 1). The node weights hi are then computed using the inverse normal CDF. The edge weights Jij = max(0, hi + hj) if two genes are connected in the network. Otherwise, Jij = 0. The magnetization 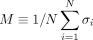 is the average state of nodes in the system. As we increase the external field H, more nodes (genes) will be active, that is, M will monotonically increase and eventually approach 1. (C–E) disease module identified by RFIM at H = −8 (point b), H = −4 (point c) and H = 8 (point d), respectively. All the active components (highlighted in red) together can be considered as the disease module.
is the average state of nodes in the system. As we increase the external field H, more nodes (genes) will be active, that is, M will monotonically increase and eventually approach 1. (C–E) disease module identified by RFIM at H = −8 (point b), H = −4 (point c) and H = 8 (point d), respectively. All the active components (highlighted in red) together can be considered as the disease module.











