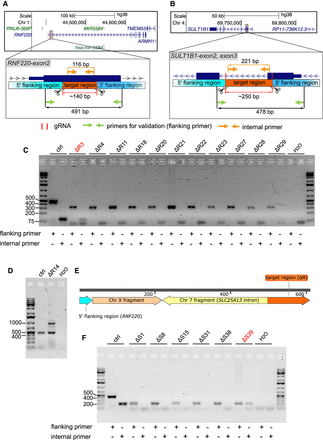
Adverse on-target genomic rearrangements occurred using the CRISPR-Cas9-gRNA delivery system. (A,B) The hg38 genome browser views (top) show the genomic locations of two protein-coding genes (RNF220 and SULT1B1). Arrows denote directionality of gene transcription. The CRISPR-Cas9-gRNA RNP targeted regions (bottom) are magnified and highlighted (orange). Cas9 cut sites (red horizontal lines and scissors), primers used for validation (flanking primers, green arrows) and for detecting target regions (internal primers, light orange arrows) are marked. The lengths of the target regions and the predicted PCR amplicons are indicated. (C) The agarose gel image distinguishes the size of PCR products generated by flanking primers to validate the deletion within the RNF220 (ΔR) gene in hTERT-RPE1 cell clones and by internal primers to detect hTERT-RPE1 clones with on-target alterations. hTERT-RPE1 clones with a confirmed deletion but also with a detectable target region are highlighted (red). Selected DNA marker bands (in bp) are depicted. (D) The agarose gel image separates the size of PCR products formed by flanking primers in a hTERT-RPE1 control (ctrl) clone (491 bp) and deletion clone ΔR14 (491 bp and ∼1000 bp). (E) The illustration depicts the composition of the extra band (∼1000 bp) in the deletion clone hTERT-RPE1 ΔR14 revealed by Sanger sequencing. The arrows show the genomic orientation and approximate size. (F) PCR-based validation in hTERT-RPE1 deletion clones as described in Figure 4C in which the SULT1B1 (ΔS) gene was modified.











