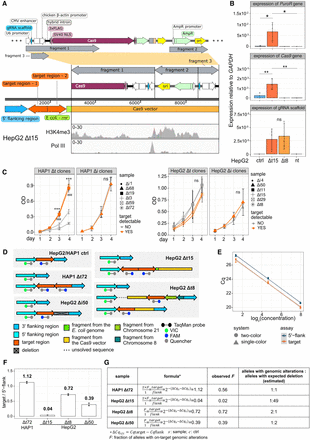
Adverse on-target genomic alterations affected cell growth, promoted active transcription, and varied in abundance among deletion clones. (A) Illustration of the different CRISPR-Cas9 vector sequences (fragment 1–3, top) that integrated in the HepG2 Δt15 deletion clone (middle). The orientation of integration is shown. Coverage tracks (bottom) show mapping of histone H3 lysine 4 trimethylation (H3K4me3) and RNA polymerase III (Pol III) ChIP-seq data to the integrated CRISPR-Cas9 vector sequences. (B) Bar graphs display expression levels of genes that integrated in the deletion clones HepG2 Δt15 and Δt8. HepG2 nontargeting clones (ctrl) as well as nontransfected (nt) cells were used as controls. Expression levels were normalized to GAPDH gene expression and determined by qPCR (n = 3, mean ± SD). Statistics: one-way ANOVA, followed by Tukey HSD test. Significance codes: (*) P < 0.05, (**) P < 0.01, ns: not significant. (C) Line graphs show the relative number of cells (measured by optical density, OD) cultured over 4 d after seeding and measured by the crystal violet assay (n = 3, mean ± SD). Statistics: one-way ANOVA followed by Tukey HSD test. Significance code: P < 0.001 (*** and ### when compared with the HAP1 Δt3 or Δt59 deletion clone, respectively); ns, not significant. (D) The design of the TaqMan assay applied in control and deletion clones is illustrated. Allelic on-target genomic alteration based on Xdrop-LRS are shown for each clone. (E) The dot plot shows the quantification cycle (Cq) using genomic template DNA from the HAP1 control clone at increasing concentrations. Both the one-color (circle) and the two-color (triangle) system were tested in the two TaqMan assays, detecting either the target region (orange) or the 5′ flanking region (dark blue). Linear models were built for each assay. (F) The bar plot displays the ratios (normalized signals) between the number of molecules containing the target and the 5′ flanking regions in the deletion clones HAP1 Δt72 and HepG2 Δt15, Δt8, Δi50. The numbers on the top of the bars display the ratio. (G) The table contains the formula used to calculate the fraction (F) of alleles carrying the intact target region from the PCR quantification cycle (Cq) and the estimated ratios between the alleles with an on-target genomic aberration and expected deletion based on the F values for each deletion clone and experiment. The cell type origin was considered when calculating ΔCqs−ΔCqc, whereby the deletion clone HAP1 Δt72 was normalized to the HAP1 control clone, whereas deletion clones HepG2 Δt15, Δt8, Δi50 were normalized to the HepG2 control clone.











