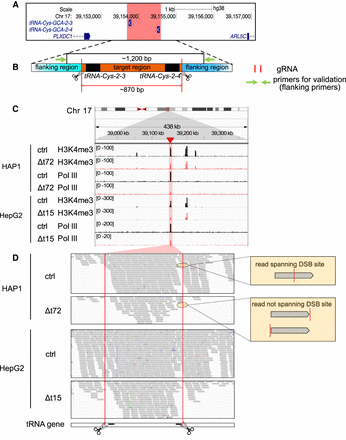
The target region remained functional in deletion clones. (A) The hg38 genome browser view shows the genomic location of our target locus (red). Arrows denote directionality of gene transcription. (B) Illustration of the design strategy of Cas9 dual guide RNA (gRNA) deletion strategy. Cas9 cut sites (red horizontal lines and scissors) and primers for validation (green arrows) are indicated. The target region (orange) containing two target transfer RNA (tRNA) genes (black) is around 870 bp. The size of the PCR product in control clones is around 1200 bp. (C) The hg38 genome browser view shows normalized ChIP-seq reads for histone H3 lysine 4 trimethylation (H3K4me3) and RNA polymerase III (Pol III) covering the target loci (highlighted in red) in the HAP1 control (ctrl) and Δt72 as well as HepG2 ctrl and Δt15 clones. (D) Alignment tracks show individual H3K4me3 ChIP-seq reads of the deleted and surrounding region as in (C). DSB sites (red lines and scissors) and tRNA gene locations (black) are indicated. Examples of reads spanning (top) or not spanning (bottom) double-strand break (DSB) site are illustrated (yellow box).











