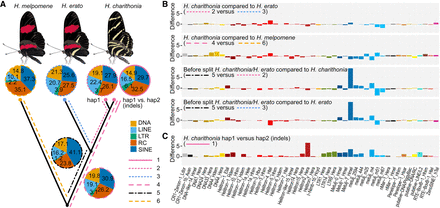
Phylogenetic dynamics of transposable elements (TEs). (A) Lineage-specific TE family accumulation. Different line types depict different branches in the phylogeny studied and allow to investigate changes in temporal accumulation of TEs. 1, TE families associated with indels between the H. charithonia haplotypes; 2, TE families accumulated in H. charithonia since the split from H. erato; 3, TE families accumulated in H. erato since the split from H. charithonia; 4, TE families accumulated in the H. erato lineage since their split from a common ancestor with H. melpomene; 5, TE families accumulated after the H. charithonia/H. erato lineage split from the common ancestor with H. melpomene but before H. erato and H. charithonia split; 6, TE families accumulated in the H. charithonia/H. erato lineage since their split from a common ancestor with H. melpomene. DNA, DNA transposons that do not involve an RNA intermediate; LINE, long interspersed nuclear elements, which encode reverse transcriptase but lack LTRs; LTR, long terminal repeats, which encode reverse transcriptase; RC, transpose by rolling-circle replication via a single-stranded DNA intermediate (Helitrons); SINE, short interspersed nuclear elements that do not encode reverse transcriptase. (B) Difference in TEs (percentage of total) between branches in the phylogeny considering the same 48 most significantly divergent TE families. Positive values indicate higher accumulation in the first branch; negative values indicate higher accumulation in the second branch of the comparison. Total TE accumulation patterns per lineage are shown in Supplemental Figure S1. (C) Difference in TEs (percentage of total) between the two H. charithonia haplotypes considering the same 48 most significantly divergent TE families.











