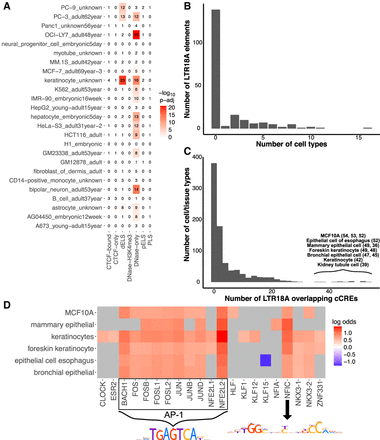
LTR18A elements are associated with enhancer epigenetic marks in human. (A) Overlap of LTR18A with ENCODE cCREs across 25 full-classification cell/tissue types. (dELS) Distal enhancer-like signature; (pELS) proximal enhancer-like signature; (PLS) promoter-like signature. The number of elements that overlap with cCREs are shown as well as their −log10-adjusted P-value by BEDTools Fisher. (B) Distribution of LTR18A elements overlapping cCREs across multiple full-classification cell/tissue types. (C) Distribution of cell/tissue types overlapping LTR18A elements. The top cell/tissue types are displayed with the number of LTR18A elements that overlap with a cCRE. (D) Motifs associated with the cCRE-overlapping LTR18A elements from the top cell/tissue types in C. Gray indicates nonsignificance at an adjusted P-value threshold of 0.05. PWMs for JUN (AP-1-related factors) and NFIC are shown.











