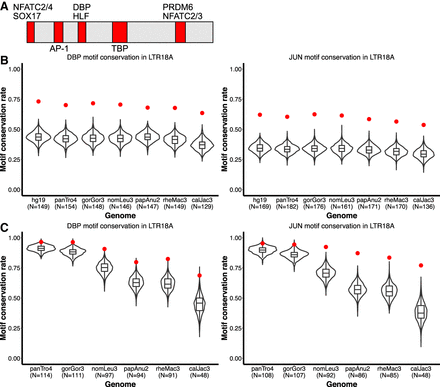
DBP and JUN motifs are more conserved than expected. (A) Motifs that are fully encompassed within shared, conserved 10-bp sliding windows across seven primate species. Motif locations in red are relative to the LTR18A Repbase consensus sequence. (B) Distribution of expected neutral DBP and JUN motif conservation rates from the consensus motif across primate species. One thousand simulations are displayed for each species. The observed conservation rate is shown by the red point. (C) Same as B, but for conservation rates from the hg19 ortholog as reference.











