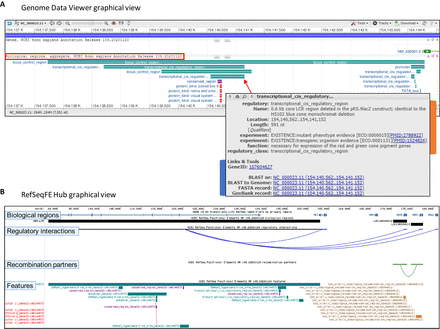
Graphical displays of RefSeqFE data. (A) NCBI Genome Data Viewer display of genome-annotated features at the human opsin locus control region (OPSIN-LCR, GeneID:107604627, also shown in Supplemental Fig. S1). Underlying features are aggregated and displayed in the “Biological regions, aggregate” track (outlined in red). Depending on user track set options or the entry point to GDV, the track may need to be turned on via the configuration interface, as detailed on our web page (Supplemental Table S1, graphical displays link). Features are color coded according to class or type. Coordinates are based on positions on the genome sequence. An example of a mouseover-activated pop-up box is shown (overlaid gray box). These boxes contain descriptive and functional information (orange tab) (Nathans et al. 1989; Wang et al. 1992), including experimental evidence and links to publications, as well as a “Links & Tools” area (blue tab) linking to the related Gene database record and to sequences and BLAST analyses. (B) RefSeqFE Hub view of parental biological regions, underlying features, and gene regulatory and recombination partner interactions in the UCSC Genome Browser. Regulatory interactions are shown between the hemoglobin subunit alpha locus control region (HBA-LCR, GeneID:106144573) and the downstream HBZ, HBA2, HBA2, and HBQ1 genes (blue curved lines), whereas the recombination partners track visualizes recombination (green curved line) between two hemoglobin subunit alpha recombination regions (LOC106804612 and LOC106804613). Parental biological regions are denoted by black rectangles in the biological regions track, and the features track uses color coding as described for A. Further item-specific metadata, display options, and links to related data and tools can be found within item- and track-specific details pages. Depending on the density of interactions in a region, appropriate zoom levels or configuration modes may need to be adjusted, or specific hub settings such as multiregion view can be used for viewing interactions between distally located regions.











