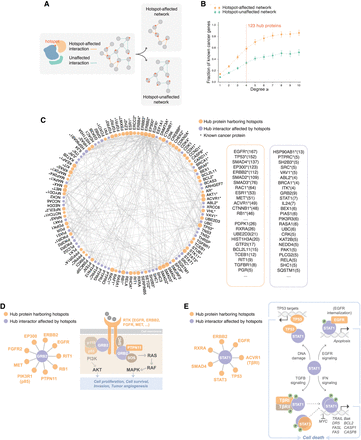
Identification of novel cancer proteins using our hotspot-affected network. (A) Schematic illustration of constructing hotspot-affected and hotspot-unaffected networks. (B) Association of proteins in the hotspot-affected and hotspot-unaffected networks with previously known cancer proteins. (C) A network view of hub proteins prioritized by our hotspot-affected network. Proteins that harbor hotspots are shown in orange, and proteins with no hotspots are shown in purple. Listed proteins are ranked first by whether the protein is a known cancer protein (indicated by asterisk) and then by their degree in the hotspot-affected network (shown in parentheses). (D,E) Implication of the top-ranked hub proteins GRB2 (D) and STAT1 (E) in oncogenesis.











