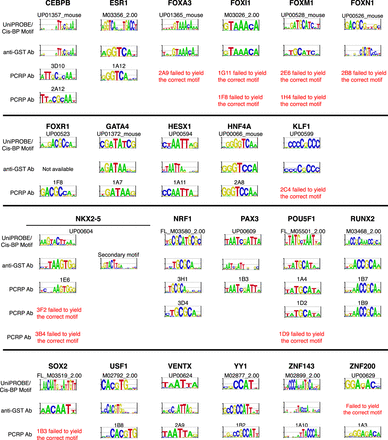
TF binding motifs derived from PBM experiments performed using anti-GST or PCRP antibodies. Each full-length TF was assayed with its PCRP mAb(s) and compared against its corresponding motif derived from an anti-GST PBM experiment and anticipated motif from the UniPROBE or Cis-BP databases (Weirauch et al. 2014; Hume et al. 2015). The TFs from UniPROBE or Cis-BP were assayed as extended DNA-binding domains. For the display of sequence motifs, probability matrices were trimmed from left and right until two consecutive positions with information content of 0.3 or greater were encountered, and logos were generated from the resulting trimmed matrices using enoLOGOS (Workman et al. 2005).











