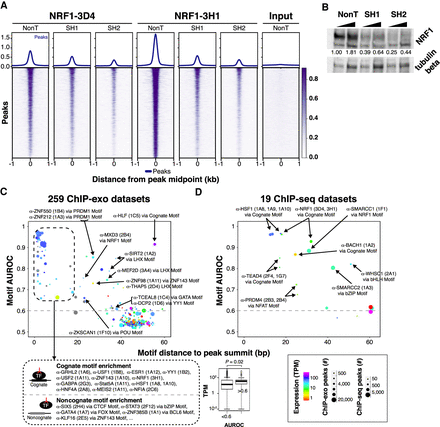
Examining specificity of PCRP mAbs. (A) Heatmaps and composite plots displaying the global loss of NRF1-3D4 and NRF1-3H1 ChIP-seq signal after NRF1 RNAi. HCT116 cells were treated with nontargeting (sh Control) or two different NRF1-directed shRNAs (shRNA 1 and shRNA 2). Rows are linked across samples and sorted in descending order by mean score per region. (B) Western blot analysis of NRF1 knockdown by two different shRNAs (SH1 and SH2) or a nontargeting shRNA (NonT). HCT116 cells were infected with the indicated shRNAs and selected with puromycin (2 μg/mL). Total cell extracts were prepared for SDS-PAGE and immunoblotting against NRF1 and tubulin beta as the loading control. NRF1 knockdown efficiency (upper band in top panel) was quantified after normalizing with tubulin beta levels using ImageJ, and the normalized values shown. (C) Motif enrichment analysis of ChIP-exo. Cartoons depict models for binding via the cognate motif of the target ssTF or noncognate binding. Box plots of TPM expression values of target ssTFs associated to antibodies stratified by AUROC value. Results from analysis of 100 putative ssTF binding motifs within each ChIP-exo data set with more than 500 peaks (259 data sets in total). We assigned to each ChIP-exo data set the PWM with the highest AUROC (“top motif”) and quantified its centering as the mean distance of the PWM match from the peak's summit. In the scatter plot, each point represents the enrichment/centering of the top motif in one of the 259 putative TF ChIP-exo data sets. Colors indicate the expression level (RNA-seq TPM value; unavailable values are shown in gray) (The ENCODE Project Consortium 2012) of the gene specific for the antibody used in the ChIP assay. Point sizes indicate the number of ChIP-exo peaks in the data set. Top motifs with AUROC > 0.6 (dashed line) and TPM values from duplicate RNA-seq experiments are indicated. (D) Results from enrichment analysis of 100 TF binding motifs within each of 19 ChIP-seq data sets. Points are formatted as in C.











