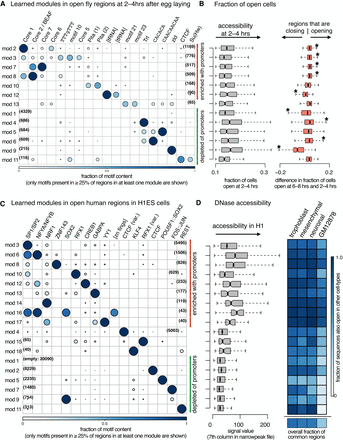
cisDIVERSITY run on open regions. (A) Thirteen modules and 28 motifs (Supplemental Fig. S13) are learned on ATAC-seq regions, which are open in at least 10% of the cells probed 2–4 h after egg laying. Only the 19 motifs that contribute to at least a quarter of the sequences in some module are shown here for clarity. Modules are reordered: The red and green modules are significantly (hypergeometric P < 10−4) enriched with promoters and depleted of them, respectively. (B) Gray indicates there are only a few differences in the fraction of cells open within each module. Orange indicates modules 2, 3, 7, 8, 4, and 6 are significantly more open in the cells 6–8 h after egg-laying, whereas modules 12, 1, and 9 are closing at that time point. (C) Eighteen modules and 21 motifs (Supplemental Fig. S14) are learned on DNase-seq regions in H1 ESCs. Again, only the motifs appearing in a quarter of sequences of some module are shown here. Red and green modules are as in A. (D) Gray indicates promoter modules have a higher DNase signal in general, but there are variations among them. Blue indicates the fraction of each module (and total below) that is also open in trophoblast, mesenchymal, and neuronal stem cells (all derived from H1 ESCs), and GM12878 shows considerable variation across modules and cell types.











