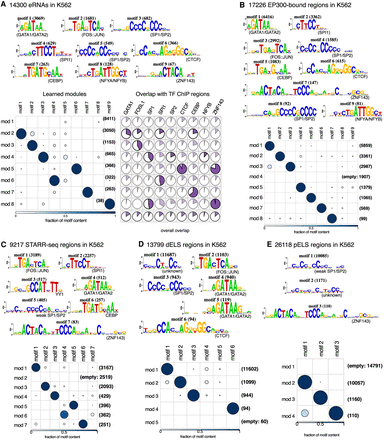
cisDIVERSITY run on putative enhancers in K562. (A) On distant eRNAs, overlap with ChIP-seq data is significant (hypergeometric P < 10−4; shown in bold) in modules that contain the matching TF motif. Note that in some cases the overlap looks large but does not show up as significant, because the hypergeometric test corrects for the sizes of the overlaps and the modules. (B) Similar motifs are found in EP300 ChIP-seq data. (C) In STARR-seq peaks, YY1 is additionally discovered. CTCF and NFYA/NFYB are not enriched. (D,E) Distant (D) and proximal enhancer-like sequences (E) deduced from chromatin signatures have fewer motif-like signatures. cisDIVERSITY run on putative enhancers in K562.











