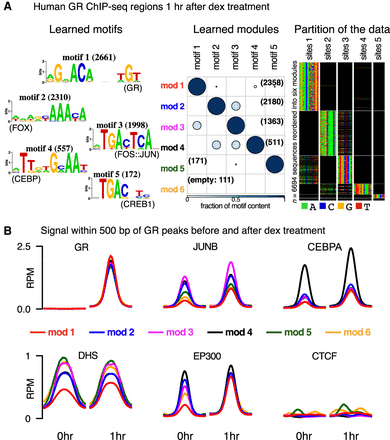
Figure 5.
Diverse signals discovered in GR-bound ChIP-seq regions. (A) cisDIVERSITY identifies six modes and five motifs in the GR ChIP-seq regions. (B) The average DNase hypersensitivity signal and input-subtracted ChIP-seq signal in reads per million (RPM) of five TFs—GR, JUNB, CEBPA, EP300, and CTCF—before and after treatment at the GR-bound regions are shown for each module.











