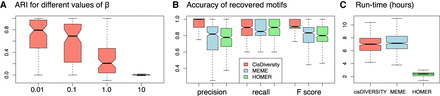
Performance on 320 simulated data sets. (A) Recovery of modules. Low values of beta result into modules with more extreme (zero or one) probability distributions of motifs. This is where cisDIVERSITY does better in recovering the planted modules. For beta = 10, the performance with respect to recovery of modules is similar to what a random clustering approach would do. (B) Recovery of motifs. Precision, recall, and F-score of recovered motifs across the 320 data sets for three different programs. (C) Time taken. All programs were run on a single core (Methods).











