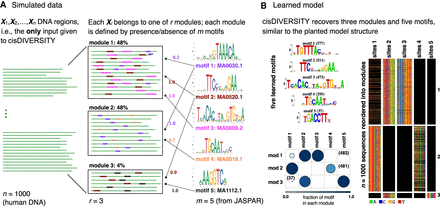
cisDIVERSITY. (A) DNA regions reported by the experiment are given as input to cisDIVERSITY. In this simulation, the n = 1000 regions are a mixture of three kinds of regions: Each region resembles one of r = 3 regulatory modules. Each module can be represented in terms of the probability of occurrence of m = 5 motifs. For example, motif 1 is present in all sequences of module 2, 20% of sequences in module 1, but not at all in module 3. In contrast, motif 4 is present only in module 2 and that, too, only in 70% of the its sequences. (B) cisDIVERSITY is run with upper bounds of r ≤ 10 and m ≤ 20. cisDIVERSITY learns the planted structure in the data set. The output has three components. First is the set of motifs that are learned, second (below) is r × m Bernoulli distributions describing the learned modules, and the third is an image matrix of the data, where each DNA sequence is a row and the sites corresponding to each motif are represented in the column. If a site is absent, those cells in the column are shown in black. cisDIVERSITY recovers the five motifs (motifs 1 and 3 are the reverse complements of the planted motifs) and the three modules to a great extent. The slight variability in the number of sites and sequences in each module is expected owing to the stochastic nature of both, the PWMs as well as the learning algorithm.











