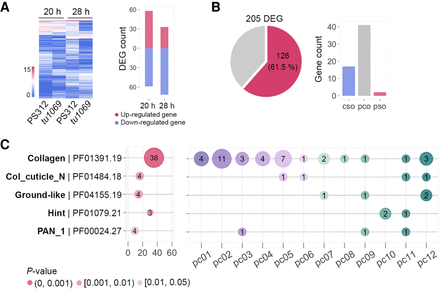
Integration of eud-1 as upstream regulator of oscillatory core genes. (A) The heatmap shows the expression levels of 205 differentially expressed genes (DEGs) between the eud-1(tu1069)-mutant and wild-type (PS312) animals at 20 h and 28 h. The bar plot indicates the number of up-/down-regulated genes. (B) The pie chart displays the overlap between DEGs and oscillating genes. The bar plot shows the number of the oscillating DEGs among the three oscillating gene classes. (C) Protein domain enrichment analysis reveals that the collagen gene family is highly enriched in the 205 DEGs, the x-axis presents the enrichment score, and the y-axis shows the name of protein domains. The size of the circles corresponds to the number of oscillatory genes in a given protein domain, and the three different colors indicate different significance levels as measured by the Fisher's exact test. The right graph profiles the overrepresented protein domains according to the 12 phase classes. The number of oscillatory genes in each phase class is marked.











