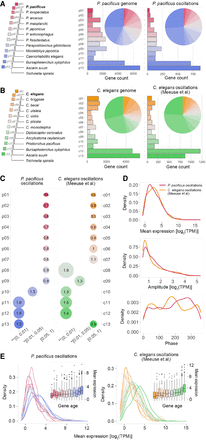
Ancient gene classes are enriched for oscillatory genes. (A) P. pacificus genes were assigned to age classes based on the most distant ortholog in a given orthology cluster. The age classes are labeled in a red (young)–blue (old) gradient. The distribution of gene number among the 13 age classes for the P. pacificus genome (n = 28,896) is shown in the central graph, and the distribution for the P. pacificus oscillating genes (n = 2964) is shown in the right part. (B) A similar analysis for the C. elegans oscillating genes set of Meeuse and coworkers (2020). (C) The graphs indicate the overrepresentation of gene age classes among the P. pacificus oscillatory genes (left) and the C. elegans oscillating genes sets of Meeuse et al. (2020). The size of the circles indicates the enrichment score; the x-axis shows the significance level measured by Fisher's exact test. (D) The plots show the distribution of average expression (top), amplitude (middle), and phase (bottom) of the oscillating genes in P. pacificus (red) and C. elegans (yellow). (E) The plots show the distribution of the mean expression of oscillating genes across 13 gene age classes in both species.











