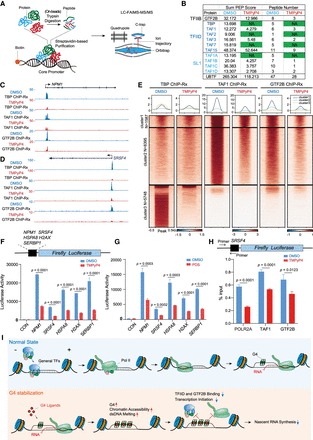
G4 stabilization inhibits transcription initiation by impairing general transcription factor loading. (A) Schematic of immunoprecipitation of G4-containing core promoter and nanoLC-FAIMS-MS/MS analysis. Streptavidin MagPoly beads were incubated with biotin-labeled G4-containing core promoter DNA and nuclear protein extract sequentially for immunoprecipitation. After on-beads trypsin digestion, the interacting proteins with core promoter DNA were analyzed on a Thermo Exploris 480 mass spectrometer. (B) List of core promoter interacting proteins with or without TMPyP4 treatment. (SL1) Selective Factor 1. Sum PEP Score is calculated as the sum of the negative logarithms of the posterior error probability (PEP) values of the connected peptide spectrum matches (PSMs). All the proteins in the list have high confidence, with q-values higher than the specified threshold 0.01. (C–E) Analysis of TBP, TAF1, and GTF2B ChIP-Rx signals in cells treated with DMSO or TMPyP4 for 2 h. Track examples at the NPM1 (C) and SRSF4 (D) loci are shown. Heat map and metaplot analyses (E) showed that TMPyP4 impaired TBP, TAF1, and GTF2B occupancy after TMPyP4 treatment. (F,G) Luciferase reporter assay of G4-containing promoters showed that G4 stabilization induced by TMPyP4 (F) or PDS (G) significantly inhibited the luciferase reporter gene expression. (H) ChIP-qPCR analysis in HEK293T cells transfected with recombinant luciferase reported plasmid. These cells were treated with or without TMPyP4. The occupancy of POLR2A, TAF1, and GTF2B in the insertion-vector junction of the recombinant plasmid was measured by ChIP-qPCR. The statistical analysis was performed using an unpaired Student's t-test. (I) Proposed model for the interplay between G4 and gene transcription. G4s are generated during transcription and mark transcription regulatory chromatin in the genome. They are correlated with active transcription and coincident with R-loops. However, disruption of G4 dynamics by G4 stabilizing ligands could alter the chromatin states by increasing the dsDNA melting and chromatin accessibility at the promoters, which impair the loading of general transcription factors, such as TBP, TFIID TAFs, and GTF2B. Thus, G4 stabilization leads to inhibition of transcription initiation and nascent RNA synthesis.











