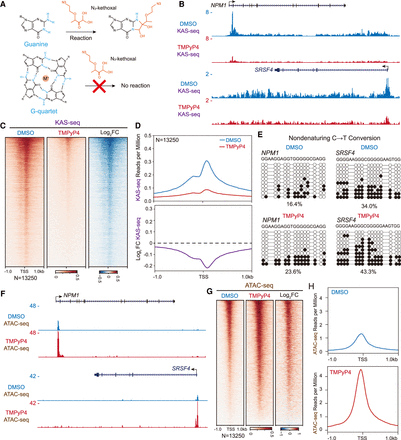
G4 stabilization by G4 ligands rapidly alters chromatin states with increased dsDNA melting and chromatin accessibility. (A) Schematic for N3-kethoxal labeling reaction in KAS-seq. In the G-quartet structure, the formation of Hoogsteen bonds blocks the chemical reaction between guanine and N3-kethoxal. (B–D) Analysis of KAS-seq signals in HEK293T cells treated with DMSO or TMPyP4. Tracks examples illustrate that TMPyP4 reduces KAS-seq signals at the promoters, gene bodies, and TESs of NPM1 and SRSF4 genes (B). Heat maps (C) and metaplots (D) demonstrate that TMPyP4 induced a global reduction of KAS-seq signals at TSSs. (E) Nondenaturing C to T conversion at two G4 forming motifs in NPM1 and SRSF4 promoters after TMPyP4 treatment in HEK293T cells. Each row shows one representative Sanger sequencing result. The converted sites are depicted as black dots, whereas the unconverted sites are represented as white dots. The percentages of C-T conversion sites for each group were calculated from these 10 sequences. (F–H) Analysis of ATAC-seq signals in the absence or presence of TMPyP4 reveals that TMPyP4 enhanced the chromatin accessibility around TSSs. Snapshots of ATAC-seq tracks at NPM1 and SRSF4 loci (F), heat maps (G), and metaplots (H) are shown.











