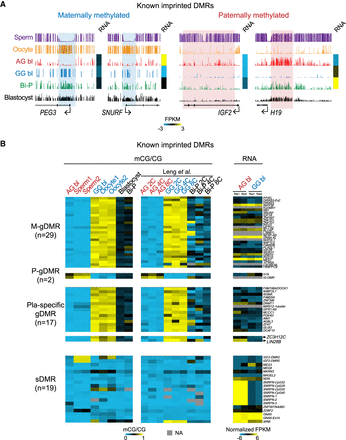
DNA methylation of known imprinted regions in the AG, GG, and Bi-P blastocysts. (A) UCSC snapshots showing the previously known DMRs located in PEG3, SNURF, IGF2, and H19. Our results showed that germline DMRs survived demethylation during early embryo development, indicating that our diploid AG and GG blastocysts are suitable models to screen for putative DMRs. The sperm and oocyte DNA methylation data are from Okae et al. (2014). (B) Heatmaps showing the DNA methylation levels of known imprinted DMRs and the related gene expression. DNA methylation in two-, four-, and eight-cell embryos published previously was included as a control (Leng et al. 2019). All replicates for the AG, GG, and Bi-P blastocysts were pooled. These known imprinted DMRs were clarified as previously reported (Okae et al. 2014). (Bi-P) Biparental embryo, (AG) androgenetic, (GG) gynogenetic.











