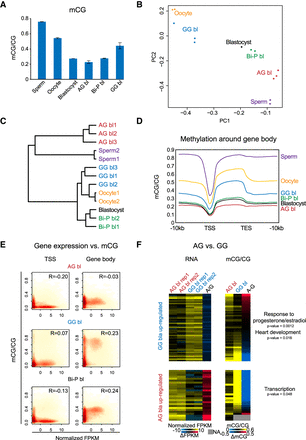
DNA methylome of diploid AG, GG, and Bi-P blastocysts. (A) Global CpG methylation in the AG, Bi-P, GG embryos, and gametes. Note that the AG blastocysts exhibited the lowest level of global DNA methylation, whereas the sperm showed the highest level, as expected. The sperm and oocyte DNA methylation data were used in the Okae et al. (2014) study. (B) Principal component analysis showed a genome-wide DNA methylation relationship among three types of blastocysts and germ cells. (C) Nonsupervised cluster analysis of the DNA methylome from different blastocysts and gametes. (D) The metaplot showing DNA methylation around gene bodies in the AG, Bi-P, GG blastocysts, and gametes. (E) The relationship between TSS (left) and gene body (right) DNA methylation and gene expression of the Bi-P blastocysts and the uniparental blastocysts. (F) Heatmaps showing DEGs in the AG and GG groups and associated gene promoter methylation levels. Only the genes expressed at one stage with FPKM ≥ 5 and at least twofold changes between two groups and with a P-value generated by DESeq2 (Love et al. 2014) less than 0.05 were selected as DEGs. (Bi-P) Biparental embryo, (AG) androgenetic, (GG) gynogenetic.











