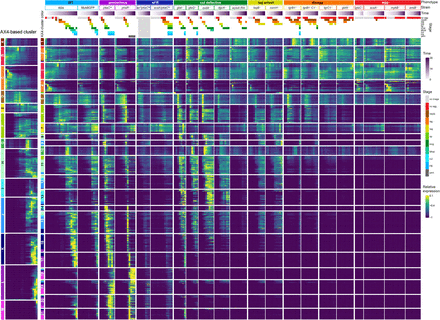
Developmental regulons are robust to temporal and genetic perturbations. We searched for genes whose expression patterns were similar despite changes in developmental time and strain genotype. The leftmost heatmap shows 13 clusters of genes that were coexpressed in the wild type AX4 during development. The clusters were assigned a letter (A–M) and a color as indicated on the left of the AX4-based column. We then reclustered these genes based on their expression patterns in all 21 strains. The resulting 21 regulons (numbered boxes with AX4-based assignment colors, left) are shown as heatmaps. Each row in the heatmaps represents a gene, each column represents a time point, and the entry colors represent relative mRNA abundances (as indicated in the relative expression scale on the right). Phenotype groups, strain names, developmental time (purple gradation, see scale on the right), and annotated morphological stages (see color legend on the right) are indicated above the heatmaps. The phenotype groups are WT (light blue), precocious development (precocious, violet), small fruiting body (sFB, dark blue), culmination defective (cul defective, dark green), tight-aggregate arrest (tag, dark yellow), tight-aggregate/loose-aggregate disaggregation (disagg, orange), and aggregationless (agg-, red). The morphological stages are no aggregation (no agg, red), rippling/stream (ripple, brown), loose aggregate (lag, orange), tight aggregate (tag, dark yellow), tipped aggregate (tip, light green), slug/first finger (slug, dark green), Mexican hat (Mhat, jade green), culmination (cul, cyan), FB (light blue), and yellow mound (yem, dark gray). Light gray indicates that no image was captured (no image, light gray).











