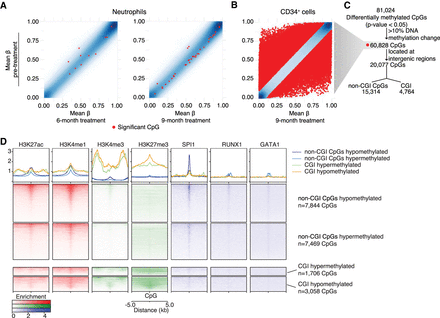
DNA methylation analysis of neutrophils and CD34+ cells CpGs. (A,B) Scatter plots show the mean of DNA methylation level (β-value) from pretreated samples (y-axis) and after 6 and 9 mo of HC treatment in neutrophils and 9 mo of HC treatment in CD34+ cells (x-axis). Red dots correspond to significantly differentially methylated CpGs according to each time point comparison. (C) Workflow of differentially methylated CpGs analyzed in CD34+ cells. All differentially methylated CpGs identified were filtered by having >10% of DNA methylation change. From these, differentially methylated CpGs overlapping intergenic regions were further subdivided according to their position in the genome in respect to CpG islands (CGI), such as differentially methylated CpGs overlapping CGIs or non-CGI CpGs. (D) Heatmap plots that show enrichment of regulatory regions (H3K27ac, H3K4me1, H3K4me3, and H3K27me3) and transcription factor bindings sites (SPI1, RUNX1, and GATA1) at differentially methylated CpGs overlapping CGIs or non-CGI CpGs using ChIP-seq data from healthy CD34+ cells.











