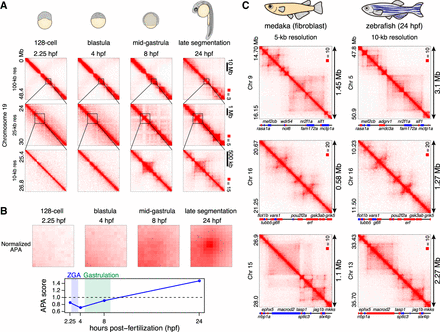
Loop establishment in zebrafish embryos. (A) Representative examples of Hi-C contact matrices of zebrafish embryos. Normalized observed Hi-C contacts are shown as a heat map at indicated resolutions. (B) Normalized APA plots are shown in the upper portion, and APA scores are shown in the lower portion. Values greater than 1 indicate the presence of loops. Hi-C contacts around loops identified in 24-hpf embryos were aggregated for each stage. (C) Examples of conserved 3D structure in synteny blocks. Hi-C maps of medaka fibroblast (5-kb resolution), and 24-hpf zebrafish embryos (10-kb resolution) are shown. The orientations of genes is indicated by different colors (forward: red; reverse: blue). Gene names are indicated for selected genes because of the space limitation.











