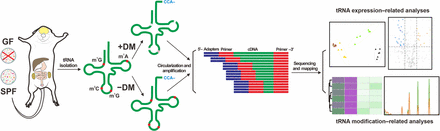
Workflow of this study. tRNAs were isolated from the total RNAs extracted from four tissue types (brain, intestine, kidney, and liver) in specific pathogen-free (SPF) and germ-free (GF) mice. Each tRNA sample was treated with (+DM) or without demethylase (−DM), respectively. After reverse transcription, circularized cDNAs were amplified and sequenced on an Illumina HiSeq system. The sequencing data were used for downstream analyses after mapping to the constructed cytosolic and mitochondrial reference tRNA data sets. (m1A) N1-methyladenosine, (m1G) N1-methylguanosine, (m3C) N3-methylcytosine.











