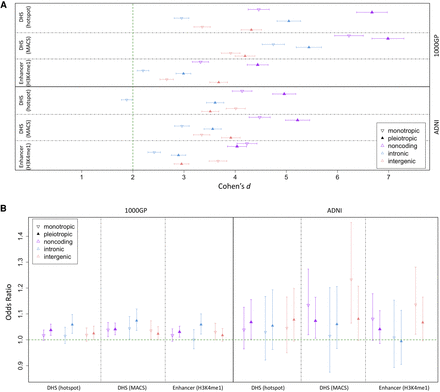
Depletion of deletions and shift of AFS overlapping DHS or enhancer sites of variable cellular activity. (A) We calculated PlyRSsum-mono (monotropic) and PlyRSsum-pleio (pleiotropic) for every deletion to quantify overlap with DHS or enhancer sites. We plot the degree of reduction in PlyRSsum-mono (or PlyRSsum-pleio) for real deletions relative to simulation measured using Cohen's d (with 95% CIs showing the uncertainty owing to the finite number of simulations). (B) For each deletion, we determined the magnitude of PlyRSsum-mono (monotropic) and PlyRSsum-pleio (pleiotropic) depletion, calculated as a ratio between its PlyRSsum-mono (or PlyRSsum-pleio) and the average PlyRSsum-mono (or PlyRSsum-pleio) of its length-matched simulated counterparts, for DHS or enhancer sites. We tested whether PlyRSsum-mono (or PlyRSsum-pleio) depletion magnitude depends on AF (deletions categorized as rare [AF ≤ 1%] or common), using multivariate logistic regression in the presence of genomic covariates. We plot the regression odds ratio with 95% profile likelihood-based CIs.











